Identification of common predisposing loci to hematopoietic cancers in four dog breeds
- PMID: 33793571
- PMCID: PMC8016107
- DOI: 10.1371/journal.pgen.1009395
Identification of common predisposing loci to hematopoietic cancers in four dog breeds
Abstract
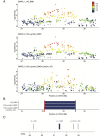
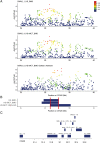
Histiocytic sarcoma (HS) is a rare but aggressive cancer in both humans and dogs. The spontaneous canine model, which has clinical, epidemiological, and histological similarities with human HS and specific breed predispositions, provides a unique opportunity to unravel the genetic basis of this cancer. In this study, we aimed to identify germline risk factors associated with the development of HS in canine-predisposed breeds. We used a methodology that combined several genome-wide association studies in a multi-breed and multi-cancer approach as well as targeted next-generation sequencing, and imputation We combined several dog breeds (Bernese mountain dogs, Rottweilers, flat-coated retrievers, and golden retrievers), and three hematopoietic cancers (HS, lymphoma, and mast cell tumor). Results showed that we not only refined the previously identified HS risk CDKN2A locus, but also identified new loci on canine chromosomes 2, 5, 14, and 20. Capture and targeted sequencing of specific loci suggested the existence of regulatory variants in non-coding regions and methylation mechanisms linked to risk haplotypes, which lead to strong cancer predisposition in specific dog breeds. We also showed that these canine cancer predisposing loci appeared to be due to the additive effect of several risk haplotypes involved in other hematopoietic cancers such as lymphoma or mast cell tumors as well. This illustrates the pleiotropic nature of these canine cancer loci as observed in human oncology, thereby reinforcing the interest of predisposed dog breeds to study cancer initiation and progression.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
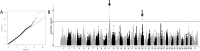

References
-
- Rao SR, Somarelli JA, Altunel E, Selmic LE, Byrum M, Sheth MU, et al. From the Clinic to the Bench and Back Again in One Dog Year: How a Cross-Species Pipeline to Identify New Treatments for Sarcoma Illuminates the Path Forward in Precision Medicine. Front Oncol. 2020;10: 117. 10.3389/fonc.2020.00117 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

