MutSα mismatch repair protein stability is governed by subunit interaction, acetylation, and ubiquitination
- PMID: 33793773
- PMCID: PMC8063085
- DOI: 10.1093/g3journal/jkaa065
MutSα mismatch repair protein stability is governed by subunit interaction, acetylation, and ubiquitination
Abstract
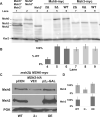
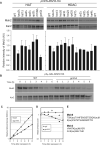
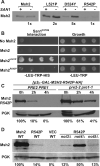
In eukaryotes, DNA mismatch recognition is accomplished by the highly conserved MutSα (Msh2/Msh6) and MutSβ (Msh2/Msh3) complexes. Previously, in the yeast Saccharomyces cerevisiae, we determined that deleting MSH6 caused wild-type Msh2 levels to drop by ∼50%. In this work, we determined that Msh6 steady-state levels are coupled to increasing or decreasing levels of Msh2. Although Msh6 and Msh2 are reciprocally regulated, Msh3 and Msh2 are not. Msh2 missense variants that are able to interact with Msh6 were destabilized when Msh6 was deleted; in contrast, variants that fail to dimerize were not further destabilized in cells lacking Msh6. In the absence of Msh6, Msh2 is turned over at a faster rate and degradation is mediated by the ubiquitin-proteasome pathway. Mutagenesis of certain conserved lysines near the dimer interface restored the levels of Msh2 in the absence of Msh6, further supporting a dimer stabilization mechanism. We identified two alternative forms of regulation both with the potential to act via lysine residues, including acetylation by Gcn5 and ubiquitination by the Not4 ligase. In the absence of Gcn5, Msh2 levels were significantly decreased; in contrast, deleting Not4 stabilized Msh2 and Msh2 missense variants with partial function. The stabilizing effect on Msh2 by either the presence of Msh6 or the absence of Not4 are dependent on Gcn5. Taken together, the results suggest that the wild-type MutSα mismatch repair protein stability is governed by subunit interaction, acetylation, and ubiquitination.
Keywords: MutS; acetylation; dimer stabilization; mismatch repair; ubiquitination.
© The Author(s) 2021. Published by Oxford University Press on behalf of Genetics Society of America.
Figures




References
-
- Amberg DC, Burke DJ, Strathern JN.. 2005. Methods in Yeast Genetics: A Cold Spring Harbor Laboratory Course Manual. Plainview, NY: Cold Spring Harbor Laboratory Press.
-
- American Cancer Society 2020. Cancer Facts & Figures 2020. Atlanta, GA: American Cancer Society.
-
- Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, et al.1989. Short Protocols in Molecular Biology. New York: John Wiley & Sons Inc.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous
