Gene dosage compensation of rRNA transcript levels in Arabidopsis thaliana lines with reduced ribosomal gene copy number
- PMID: 33793816
- PMCID: PMC8225240
- DOI: 10.1093/plcell/koab020
Gene dosage compensation of rRNA transcript levels in Arabidopsis thaliana lines with reduced ribosomal gene copy number
Abstract
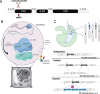
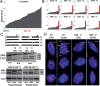
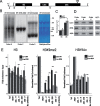
The 45S rRNA genes (rDNA) are among the largest repetitive elements in eukaryotic genomes. rDNA consists of tandem arrays of rRNA genes, many of which are transcriptionally silenced. Silent rDNA repeats may act as 'back-up' copies for ribosome biogenesis and have nuclear organization roles. Through Cas9-mediated genome editing in the Arabidopsis thaliana female gametophyte, we reduced 45S rDNA copy number (CN) to a plateau of ∼10%. Two independent lines had rDNA CNs reduced by up to 90% at the T7 generation, named low copy number (LCN) lines. Despite drastic reduction of rDNA copies, rRNA transcriptional rates, and steady-state levels remained the same as wild-type plants. Gene dosage compensation of rRNA transcript levels was associated with reduction of silencing histone marks at rDNA loci and altered Nucleolar Organiser Region 2 organization. Although overall genome integrity of LCN lines appears unaffected, a chromosome segmental duplication occurred in one of the lines. Transcriptome analysis of LCN seedlings identified several shared dysregulated genes and pathways in both independent lines. Cas9 genome editing of rRNA repeats to generate LCN lines provides a powerful technique to elucidate rDNA dosage compensation mechanisms and impacts of low rDNA CN on genome stability, development, and cellular processes.
� The Author(s) 2021. Published by Oxford University Press on behalf of American Society of Plant Biologists.
Figures


References
-
- Beying N, Schmidt C, Pacher M, Houben A, Puchta H (2020) CRISPR–Cas9-mediated induction of heritable chromosomal translocations in Arabidopsis. Nat Plants 6: 638–645 - PubMed
-
- Bray NL, Pimentel H, Melsted P, Pachter L (2016) Near-optimal probabilistic RNA-seq quantification. Nat Biotechnol 34: 525–527 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

