Enhanced nucleotide analysis enables the quantification of deoxynucleotides in plants and algae revealing connections between nucleoside and deoxynucleoside metabolism
- PMID: 33793855
- PMCID: PMC8136904
- DOI: 10.1093/plcell/koaa028
Enhanced nucleotide analysis enables the quantification of deoxynucleotides in plants and algae revealing connections between nucleoside and deoxynucleoside metabolism
Abstract
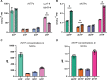
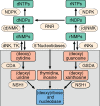
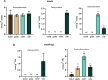
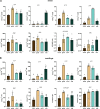
Detecting and quantifying low-abundance (deoxy)ribonucleotides and (deoxy)ribonucleosides in plants remains difficult; this is a major roadblock for the investigation of plant nucleotide (NT) metabolism. Here, we present a method that overcomes this limitation, allowing the detection of all deoxy- and ribonucleotides as well as the corresponding nucleosides from the same plant sample. The method is characterized by high sensitivity and robustness enabling the reproducible detection and absolute quantification of these metabolites even if they are of low abundance. Employing the new method, we analyzed Arabidopsis thaliana null mutants of CYTIDINE DEAMINASE, GUANOSINE DEAMINASE, and NUCLEOSIDE HYDROLASE 1, demonstrating that the deoxyribonucleotide (dNT) metabolism is intricately interwoven with the catabolism of ribonucleosides (rNs). In addition, we discovered a function of rN catabolic enzymes in the degradation of deoxyribonucleosides in vivo. We also determined the concentrations of dNTs in several mono- and dicotyledonous plants, a bryophyte, and three algae, revealing a correlation of GC to AT dNT ratios with genomic GC contents. This suggests a link between the genome and the metabolome previously discussed but not experimentally addressed. Together, these findings demonstrate the potential of this new method to provide insight into plant NT metabolism.
© American Society of Plant Biologists 2020. All rights reserved. For permissions, please email: journals.permissions@oup.com.
Figures





References
-
- Aoyagi S, Sugiyama M, Fukuda H (1998) BEN1 and ZEN1 cDNAs encoding S1-type DNases that are associated with programmed cell death in plants. FEBS Lett 429: 134–138 - PubMed
-
- Ashihara H, Crozier A, Ludwig IA (2020) Plant Nucleotide Metabolism: Biosynthesis, Degradation, and Alkaloid Formation. Wiley Blackwell, Chichester
-
- Ashihara H, Mitsui K, Ukaji T (1987) A simple analysis of purine and pyrimidine nucleotides in plant cells by high-performance liquid chromatography. Z Naturforsch C 42: 297–299
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

