Function of membrane domains in rho-of-plant signaling
- PMID: 33793925
- PMCID: PMC8133555
- DOI: 10.1093/plphys/kiaa082
Function of membrane domains in rho-of-plant signaling
Abstract
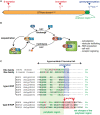
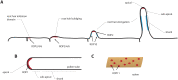
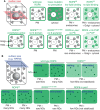
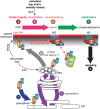
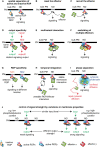
In a crowded environment, establishing interactions between different molecular partners can take a long time. Biological membranes have solved this issue, as they simultaneously are fluid and possess compartmentalized domains. This nanoscale organization of the membrane is often based on weak, local, and multivalent interactions between lipids and proteins. However, from local interactions at the nanoscale, different functional properties emerge at the higher scale, and these are critical to regulate and integrate cellular signaling. Rho of Plant (ROP) proteins are small guanosine triphosphate hydrolase enzymes (GTPases) involved in hormonal, biotic, and abiotic signaling, as well as fundamental cell biological properties such as polarity, vesicular trafficking, and cytoskeleton dynamics. Association with the membrane is essential for ROP function, as well as their precise targeting within micrometer-sized polar domains (i.e. microdomains) and nanometer-sized clusters (i.e. nanodomains). Here, we review our current knowledge about the formation and the maintenance of the ROP domains in membranes. Furthermore, we propose a model for ROP membrane targeting and discuss how the nanoscale organization of ROPs in membranes could determine signaling parameters like signal specificity, amplification, and integration.
© The Author(s) 2021. Published by Oxford University Press on behalf of American Society of Plant Biologists.
Figures

References
-
- Barbosa ICR, Rojas-Murcia N, Geldner N (2019) The Casparian strip—one ring to bring cell biology to lignification? Curr Opin Biotechnol 56:121–129 - PubMed
-
- Berken A, Wittinghofer A (2008) Structure and function of Rho-type molecular switches in plants. Plant Physiol Biochem PPB 46:380–393 - PubMed
-
- Bourne HR, Sanders DA, McCormick F (1991) The GTPase superfamily: conserved structure and molecular mechanism. Nature 349:117–127 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

