Revising Berg-Purcell for finite receptor kinetics
- PMID: 33794148
- PMCID: PMC8390789
- DOI: 10.1016/j.bpj.2021.03.021
Revising Berg-Purcell for finite receptor kinetics
Abstract
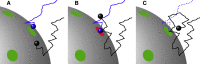
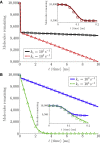
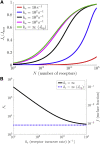
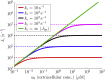
From nutrient uptake to chemoreception to synaptic transmission, many systems in cell biology depend on molecules diffusing and binding to membrane receptors. Mathematical analysis of such systems often neglects the fact that receptors process molecules at finite kinetic rates. A key example is the celebrated formula of Berg and Purcell for the rate that cell surface receptors capture extracellular molecules. Indeed, this influential result is only valid if receptors transport molecules through the cell wall at a rate much faster than molecules arrive at receptors. From a mathematical perspective, ignoring receptor kinetics is convenient because it makes the diffusing molecules independent. In contrast, including receptor kinetics introduces correlations between the diffusing molecules because, for example, bound receptors may be temporarily blocked from binding additional molecules. In this work, we present a modeling framework for coupling bulk diffusion to surface receptors with finite kinetic rates. The framework uses boundary homogenization to couple the diffusion equation to nonlinear ordinary differential equations on the boundary. We use this framework to derive an explicit formula for the cellular uptake rate and show that the analysis of Berg and Purcell significantly overestimates uptake in some typical biophysical scenarios. We confirm our analysis by numerical simulations of a many-particle stochastic system.
Copyright © 2021 Biophysical Society. Published by Elsevier Inc. All rights reserved.
Figures
Similar articles
-
The Berg-Purcell limit revisited.Biophys J. 2014 Feb 18;106(4):976-85. doi: 10.1016/j.bpj.2013.12.030. Biophys J. 2014. PMID: 24560000 Free PMC article.
-
Reaction kinetics of membrane receptors: a spatial modeling approach.J Math Biol. 2025 May 6;90(6):57. doi: 10.1007/s00285-025-02217-w. J Math Biol. 2025. PMID: 40327118
-
Effect of ligand diffusion on occupancy fluctuations of cell-surface receptors.J Chem Phys. 2013 Sep 28;139(12):121910. doi: 10.1063/1.4816105. J Chem Phys. 2013. PMID: 24089722 Free PMC article.
-
Kinetics and mechanics of cell adhesion.J Biomech. 2000 Jan;33(1):23-33. doi: 10.1016/s0021-9290(99)00163-3. J Biomech. 2000. PMID: 10609515 Review.
-
Mathematical Models of Diffusion in Physiology.Physiol Res. 2024 Aug 30;73(S1):S471-S476. doi: 10.33549/physiolres.935292. Epub 2024 Apr 22. Physiol Res. 2024. PMID: 38647169 Free PMC article. Review.
Cited by
-
Short-time diffusive fluxes over membrane receptors yields the direction of a signalling source.R Soc Open Sci. 2023 Apr 26;10(4):221619. doi: 10.1098/rsos.221619. eCollection 2023 Apr. R Soc Open Sci. 2023. PMID: 37122946 Free PMC article.
-
Single-cell directional sensing from just a few receptor binding events.Biophys J. 2023 Aug 8;122(15):3108-3116. doi: 10.1016/j.bpj.2023.06.015. Epub 2023 Jun 24. Biophys J. 2023. PMID: 37355773 Free PMC article.
-
Extending Mathematical Frameworks to Investigate Neuronal Dynamics in the Presence of Microglial Ensheathment.Bull Math Biol. 2025 Apr 4;87(5):63. doi: 10.1007/s11538-025-01438-w. Bull Math Biol. 2025. PMID: 40183855 Free PMC article.
-
Boundary Homogenization and Numerical Modeling of Solute Transport Across the Blood-Brain Barrier.bioRxiv [Preprint]. 2025 Jul 3:2025.07.01.662658. doi: 10.1101/2025.07.01.662658. bioRxiv. 2025. PMID: 40631232 Free PMC article. Preprint.
References
-
- Johnston M. Feasting, fasting and fermenting. Glucose sensing in yeast and other cells. Trends Genet. 1999;15:29–33. - PubMed
-
- Perelson A.S., Weisbuch G. Immunology for physicists. Rev. Mod. Phys. 1997;69:1219–1268.
-
- Deutch A. Fundamental Neuroscience. Fourth Edition. Elsevier; 2013. Chapter 6; pp. 117–138.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

