Pervasive cis effects of variation in copy number of large tandem repeats on local DNA methylation and gene expression
- PMID: 33794196
- PMCID: PMC8206010
- DOI: 10.1016/j.ajhg.2021.03.016
Pervasive cis effects of variation in copy number of large tandem repeats on local DNA methylation and gene expression
Abstract
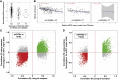
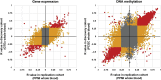
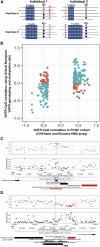
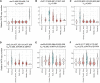
Variable number tandem repeats (VNTRs) are composed of large tandemly repeated motifs, many of which are highly polymorphic in copy number. However, because of their large size and repetitive nature, they remain poorly studied. To investigate the regulatory potential of VNTRs, we used read-depth data from Illumina whole-genome sequencing to perform association analysis between copy number of ∼70,000 VNTRs (motif size ≥ 10 bp) with both gene expression (404 samples in 48 tissues) and DNA methylation (235 samples in peripheral blood), identifying thousands of VNTRs that are associated with local gene expression (eVNTRs) and DNA methylation levels (mVNTRs). Using an independent cohort, we validated 73%-80% of signals observed in the two discovery cohorts, while allelic analysis of VNTR length and CpG methylation in 30 Oxford Nanopore genomes gave additional support for mVNTR loci, thus providing robust evidence to support that these represent genuine associations. Further, conditional analysis indicated that many eVNTRs and mVNTRs act as QTLs independently of other local variation. We also observed strong enrichments of eVNTRs and mVNTRs for regulatory features such as enhancers and promoters. Using the Human Genome Diversity Panel, we define sets of VNTRs that show highly divergent copy numbers among human populations and show that these are enriched for regulatory effects and preferentially associate with genes that have been linked with human phenotypes through GWASs. Our study provides strong evidence supporting functional variation at thousands of VNTRs and defines candidate sets of VNTRs, copy number variation of which potentially plays a role in numerous human phenotypes.
Keywords: VNTR; eQTL; mQTL; macrosatellite; minisatellite.
Copyright © 2021 American Society of Human Genetics. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
The authors declare no competing interests.
Figures


References
-
- Lander E.S., Linton L.M., Birren B., Nusbaum C., Zody M.C., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W., International Human Genome Sequencing Consortium Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

