Human OMICs and Computational Biology Research in Africa: Current Challenges and Prospects
- PMID: 33794662
- PMCID: PMC8060717
- DOI: 10.1089/omi.2021.0004
Human OMICs and Computational Biology Research in Africa: Current Challenges and Prospects
Abstract
Following the publication of the first human genome, OMICs research, including genomics, transcriptomics, proteomics, and metagenomics, has been on the rise. OMICs studies revealed the complex genetic diversity among human populations and challenged our understandings of genotype-phenotype correlations. Africa, being the cradle of the first modern humans, is distinguished by a large genetic diversity within its populations and rich ethnolinguistic history. However, the available human OMICs tools and databases are not representative of this diversity, therefore creating significant gaps in biomedical research. African scientists, students, and publics are among the key contributors to OMICs systems science. This expert review examines the pressing issues in human OMICs research, education, and development in Africa, as seen through a lens of computational biology, public health relevant technology innovation, critically-informed science governance, and how best to harness OMICs data to benefit health and societies in Africa and beyond. We underscore the disparities between North and Sub-Saharan Africa at different levels. A harmonized African ethnolinguistic classification would help address annotation challenges associated with population diversity. Finally, building on the existing strategic research initiatives, such as the H3Africa and H3ABioNet Consortia, we highly recommend addressing large-scale multidisciplinary research challenges, strengthening research collaborations and knowledge transfer, and enhancing the ability of African researchers to influence and shape national and international research, policy, and funding agendas. This article and analysis contribute to a deeper understanding of past and current challenges in the African OMICs innovation ecosystem, while also offering foresight on future innovation trajectories.
Keywords: Africa; African OMICs innovation ecosystem; bioinformatics; computational biology; diversity; foresight; public health.
Conflict of interest statement
No competing financial interests exist.
Figures

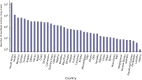


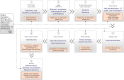

References
-
- Abimiku A, Mayne ES, Joloba M, Beiswanger CM, Troyer J, and Wideroff L. (2017). H3Africa Biorepository Program: Supporting genomics research on African populations by sharing high-quality biospecimens. Biopreser Biobank 15, 99–102
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

