Identification of the angiogenesis related genes for predicting prognosis of patients with gastric cancer
- PMID: 33794777
- PMCID: PMC8017607
- DOI: 10.1186/s12876-021-01734-4
Identification of the angiogenesis related genes for predicting prognosis of patients with gastric cancer
Abstract
Introduction: Angiogenesis is a key factor in promoting tumor growth, invasion and metastasis. In this study we aimed to investigate the prognostic value of angiogenesis-related genes (ARGs) in gastric cancer (GC).
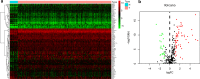
Methods: mRNA sequencing data with clinical information of GC were downloaded from The Cancer Genome Atlas (TCGA) and the Gene Expression Omnibus (GEO) databases. The differentially expressed ARGs between normal and tumor tissues were analyzed by limma package, and then prognosis‑associated genes were screened using Cox regression analysis. Nine angiogenesis genes were identified as crucially related to the overall survival (OS) of patients through least absolute shrinkage and selection operator (LASSO) regression. The prognostic model and corresponding nomograms were establish based on 9 ARGs and verified in in both TCGA and GEO GC cohorts respectively.
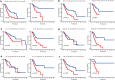
Results: Eighty-five differentially expressed ARGs and their enriched pathways were confirmed. Significant enrichment analysis revealed that ARGs-related signaling pathway genes were highly related to tumor angiogenesis development. Kaplan-Meier analysis revealed that patients in the high-risk group had worse OS rates compared with the low-risk group in training cohort and validation cohort. In addition, RS had a good prognostic effect on GC patients with different clinical features, especially those with advanced GC. Besides, the calibration curves verified fine concordance between the nomogram prediction model and actual observation.
Conclusions: We developed a nine gene signature related to the angiogenesis that can predict overall survival for GC. It's assumed to be a valuable prognosis model with high efficiency, providing new perspectives in targeted therapy.
Keywords: Angiogenesis; Gastric cancer; Gene; Prognostic.
Conflict of interest statement
None of the authors has any conflict of interest to disclose.
Figures





References
-
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

