Incorporation of a nucleoside analog maps genome repair sites in postmitotic human neurons
- PMID: 33795458
- PMCID: PMC9179101
- DOI: 10.1126/science.abb9032
Incorporation of a nucleoside analog maps genome repair sites in postmitotic human neurons
Abstract
Neurons are the longest-lived cells in our bodies and lack DNA replication, which makes them reliant on a limited repertoire of DNA repair mechanisms to maintain genome fidelity. These repair mechanisms decline with age, but we have limited knowledge of how genome instability emerges and what strategies neurons and other long-lived cells may have evolved to protect their genomes over the human life span. A targeted sequencing approach in human embryonic stem cell-induced neurons shows that, in neurons, DNA repair is enriched at well-defined hotspots that protect essential genes. These hotspots are enriched with histone H2A isoforms and RNA binding proteins and are associated with evolutionarily conserved elements of the human genome. These findings provide a basis for understanding genome integrity as it relates to aging and disease in the nervous system.
Copyright © 2021 The Authors, some rights reserved; exclusive licensee American Association for the Advancement of Science. No claim to original U.S. Government Works.
Figures
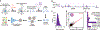
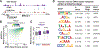

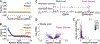
Comment in
-
Neuronal enhancers get a break.Neuron. 2021 Jun 2;109(11):1766-1768. doi: 10.1016/j.neuron.2021.05.008. Neuron. 2021. PMID: 34081919
-
Mapping the hotspots for DNA repair synthesis in human brain organoids.Cell Death Differ. 2021 Nov;28(11):3193-3195. doi: 10.1038/s41418-021-00863-3. Epub 2021 Aug 31. Cell Death Differ. 2021. PMID: 34465892 Free PMC article. No abstract available.
References
-
- Chow HM, Herrup K, Nat. Rev. Neurosci 16, 672–684 (2015). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

