The virome of German bats: comparing virus discovery approaches
- PMID: 33795699
- PMCID: PMC8016945
- DOI: 10.1038/s41598-021-86435-4
The virome of German bats: comparing virus discovery approaches
Abstract
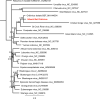
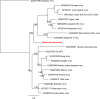
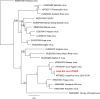
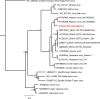
Bats are known to be reservoirs of several highly pathogenic viruses. Hence, the interest in bat virus discovery has been increasing rapidly over the last decade. So far, most studies have focused on a single type of virus detection method, either PCR, virus isolation or virome sequencing. Here we present a comprehensive approach in virus discovery, using all three discovery methods on samples from the same bats. By family-specific PCR screening we found sequences of paramyxoviruses, adenoviruses, herpesviruses and one coronavirus. By cell culture we isolated a novel bat adenovirus and bat orthoreovirus. Virome sequencing revealed viral sequences of ten different virus families and orders: three bat nairoviruses, three phenuiviruses, one orbivirus, one rotavirus, one orthoreovirus, one mononegavirus, five parvoviruses, seven picornaviruses, three retroviruses, one totivirus and two thymoviruses were discovered. Of all viruses identified by family-specific PCR in the original samples, none was found by metagenomic sequencing. Vice versa, none of the viruses found by the metagenomic virome approach was detected by family-specific PCRs targeting the same family. The discrepancy of detected viruses by different detection approaches suggests that a combined approach using different detection methods is necessary for virus discovery studies.
Conflict of interest statement
The authors declare no competing interests.
Figures




References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

