A cell-specific regulatory region of the human ABO blood group gene regulates the neighborhood gene encoding odorant binding protein 2B
- PMID: 33795748
- PMCID: PMC8016878
- DOI: 10.1038/s41598-021-86843-6
A cell-specific regulatory region of the human ABO blood group gene regulates the neighborhood gene encoding odorant binding protein 2B
Abstract
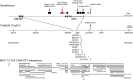
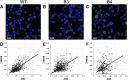
The human ABO blood group system is of great importance in blood transfusion and organ transplantation. ABO transcription is known to be regulated by a constitutive promoter in a CpG island and regions for regulation of cell-specific expression such as the downstream + 22.6-kb site for epithelial cells and a site in intron 1 for erythroid cells. Here we investigated whether the + 22.6-kb site might play a role in transcriptional regulation of the gene encoding odorant binding protein 2B (OBP2B), which is located on the centromere side 43.4 kb from the + 22.6-kb site. In the gastric cancer cell line KATOIII, quantitative PCR analysis demonstrated significantly reduced amounts of OBP2B and ABO transcripts in mutant cells with biallelic deletions of the site created using the CRISPR/Cas9 system, relative to those in the wild-type cells, and Western blotting demonstrated a corresponding reduction of OBP2B protein in the mutant cells. Moreover, single-molecule fluorescence in situ hybridization assays indicated that the amounts of both transcripts were correlated in individual cells. These findings suggest that OBP2B could be co-regulated by the + 22.6-kb site of ABO.
Conflict of interest statement
The authors declare no competing interests.
Figures




References
-
- Daniels, G. Human Blood Groups (ed. Daniels, G.). 21–25 (Wiley-Blackwell, 2013).
-
- Yamamoto F. Molecular genetics of ABO. Vox Sang. 2000;78:91–103. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

