Adhering interacting cells to two opposing coverslips allows super-resolution imaging of cell-cell interfaces
- PMID: 33795833
- PMCID: PMC8016881
- DOI: 10.1038/s42003-021-01960-2
Adhering interacting cells to two opposing coverslips allows super-resolution imaging of cell-cell interfaces
Abstract
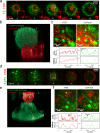
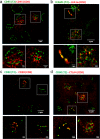
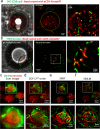
Cell-cell interfaces convey mechanical and chemical information in multicellular systems. Microscopy has revealed intricate structure of such interfaces, yet typically with limited resolution due to diffraction and unfavourable orthogonal orientation of the interface to the coverslip. We present a simple and robust way to align cell-cell interfaces in parallel to the coverslip by adhering the interacting cells to two opposing coverslips. We demonstrate high-quality diffraction-limited and super-resolution imaging of interfaces (immune-synapses) between fixed and live CD8+ T-cells and either antigen presenting cells or melanoma cells. Imaging methods include bright-field, confocal, STED, dSTORM, SOFI, SRRF and large-scale tiled images. The low background, lack of aberrations and enhanced spatial stability of our method relative to existing cell-trapping techniques allow use of these methods. We expect that the simplicity and wide-compatibility of our approach will allow its wide dissemination for super-resolving the intricate structure and molecular organization in a variety of cell-cell interfaces.
Conflict of interest statement
The authors declare no competing interests.
Figures


References
-
- Cooper, G. M. The Cell, A molecular approach (Sinauer Associates, 2000).
-
- Nicholls, J. G. From neuron to brain 5th edn, (Sinauer Associates, 2012).
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

