A PRC2-independent function for EZH2 in regulating rRNA 2'-O methylation and IRES-dependent translation
- PMID: 33795875
- PMCID: PMC8162121
- DOI: 10.1038/s41556-021-00653-6
A PRC2-independent function for EZH2 in regulating rRNA 2'-O methylation and IRES-dependent translation
Abstract
Dysregulated translation is a common feature of cancer. Uncovering its governing factors and underlying mechanism are important for cancer therapy. Here, we report that enhancer of zeste homologue 2 (EZH2), previously known as a transcription repressor and lysine methyltransferase, can directly interact with fibrillarin (FBL) to exert its role in translational regulation. We demonstrate that EZH2 enhances rRNA 2'-O methylation via its direct interaction with FBL. Mechanistically, EZH2 strengthens the FBL-NOP56 interaction and facilitates the assembly of box C/D small nucleolar ribonucleoprotein. Strikingly, EZH2 deficiency impairs the translation process globally and reduces internal ribosome entry site (IRES)-dependent translation initiation in cancer cells. Our findings reveal a previously unrecognized role of EZH2 in cancer-related translational regulation.
Conflict of interest statement
Competing interests
No potential conflict of interest to disclose.
Figures
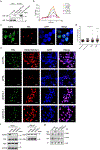






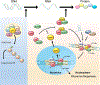



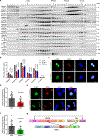


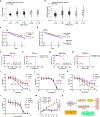
References
-
- Pelletier J, Thomas G & Volarevic S Ribosome biogenesis in cancer: new players and therapeutic avenues. Nature reviews. Cancer 18, 51–63 (2018). - PubMed
-
- Sharma S & Lafontaine DLJ ‘View From A Bridge’: A New Perspective on Eukaryotic rRNA Base Modification. Trends in biochemical sciences 40, 560–575 (2015). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

