Detection of respiratory syncytial virus defective genomes in nasal secretions is associated with distinct clinical outcomes
- PMID: 33795879
- PMCID: PMC9098209
- DOI: 10.1038/s41564-021-00882-3
Detection of respiratory syncytial virus defective genomes in nasal secretions is associated with distinct clinical outcomes
Abstract
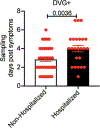
Respiratory syncytial virus (RSV) causes respiratory illness in children, immunosuppressed individuals and the elderly. However, the viral factors influencing the clinical outcome of RSV infections remain poorly defined. Defective viral genomes (DVGs) can suppress virus replication by competing for viral proteins and by stimulating antiviral immunity. We studied the association between detection of DVGs of the copy-back type and disease severity in three RSV A-confirmed cohorts. In hospitalized children, detection of DVGs in respiratory samples at or around the time of admission associated strongly with more severe disease, higher viral load and a stronger pro-inflammatory response. Interestingly, in experimentally infected adults, the presence of DVGs in respiratory secretions differentially associated with RSV disease severity depending on when DVGs were detected. Detection of DVGs early after infection associated with low viral loads and mild disease, whereas detection of DVGs late after infection, especially if DVGs were present for prolonged periods, associated with high viral loads and severe disease. Taken together, we demonstrate that the kinetics of DVG accumulation and duration could predict clinical outcome of RSV A infection in humans, and thus could be used as a prognostic tool to identify patients at risk of worse clinical disease.
Figures



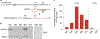
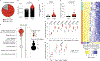

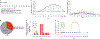
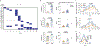

References
-
- Falsey AR, Hennessey PA, Formica MA, Cox C & Walsh EE Respiratory syncytial virus infection in elderly and high-risk adults. N. Engl. J. Med. 352, 1749–1759 (2005). - PubMed
-
- Meissner HC et al. Immunoprophylaxis with palivizumab, a humanized respiratory syncytial virus monoclonal antibody, for prevention of respiratory syncytial virus infection in high risk infants: a consensus opinion. Pediatr. Infect. Dis. J. 18, 223–231 (1999). - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

