Gain of toxic function by long-term AAV9-mediated SMN overexpression in the sensorimotor circuit
- PMID: 33795885
- PMCID: PMC8254787
- DOI: 10.1038/s41593-021-00827-3
Gain of toxic function by long-term AAV9-mediated SMN overexpression in the sensorimotor circuit
Abstract
The neurodegenerative disease spinal muscular atrophy (SMA) is caused by deficiency in the survival motor neuron (SMN) protein. Currently approved SMA treatments aim to restore SMN, but the potential for SMN expression beyond physiological levels is a unique feature of adeno-associated virus serotype 9 (AAV9)-SMN gene therapy. Here, we show that long-term AAV9-mediated SMN overexpression in mouse models induces dose-dependent, late-onset motor dysfunction associated with loss of proprioceptive synapses and neurodegeneration. Mechanistically, aggregation of overexpressed SMN in the cytoplasm of motor circuit neurons sequesters components of small nuclear ribonucleoproteins, leading to splicing dysregulation and widespread transcriptome abnormalities with prominent signatures of neuroinflammation and the innate immune response. Thus, long-term SMN overexpression interferes with RNA regulation and triggers SMA-like pathogenic events through toxic gain-of-function mechanisms. These unanticipated, SMN-dependent and neuron-specific liabilities warrant caution on the long-term safety of treating individuals with SMA with AAV9-SMN and the risks of uncontrolled protein expression by gene therapy.
Conflict of interest statement
Competing Interests
The authors declare no competing interests.
Figures

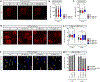
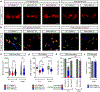

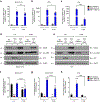
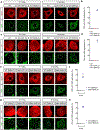
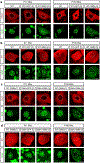
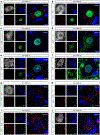

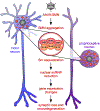
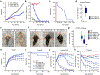


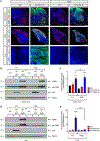
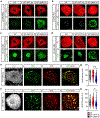
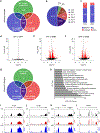
Comment in
-
Maybe too much of a good thing in gene therapy.Nat Neurosci. 2021 Jul;24(7):901-902. doi: 10.1038/s41593-021-00882-w. Nat Neurosci. 2021. PMID: 34131327 No abstract available.
-
Assuring long-term safety of highly effective gene-modulating therapeutics for rare diseases.J Clin Invest. 2021 Aug 2;131(15):e152817. doi: 10.1172/JCI152817. J Clin Invest. 2021. PMID: 34338226 Free PMC article. No abstract available.
-
Commentary: Current Status of Gene Therapy for Spinal Muscular Atrophy.Front Cell Neurosci. 2022 May 17;16:916065. doi: 10.3389/fncel.2022.916065. eCollection 2022. Front Cell Neurosci. 2022. PMID: 35656408 Free PMC article. No abstract available.
References
-
- Lefebvre S et al. Identification and characterization of a spinal muscular atrophy-determining gene. Cell 80, 155–65 (1995). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

