Improved Detection of HIV Gag p24 Protein Using a Combined Immunoprecipitation and Digital ELISA Method
- PMID: 33796087
- PMCID: PMC8007784
- DOI: 10.3389/fmicb.2021.636703
Improved Detection of HIV Gag p24 Protein Using a Combined Immunoprecipitation and Digital ELISA Method
Abstract
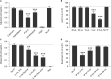
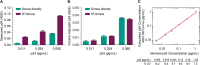
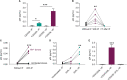
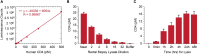
Greater than 90% of HIV-1 proviruses are thought to be defective and incapable of viral replication. While replication competent proviruses are of primary concern with respect to disease progression or transmission, studies have shown that even defective proviruses are not silent and can produce viral proteins, which may contribute to inflammation and immune responses. Viral protein expression also has implications for immune-based HIV-1 clearance strategies, which rely on antigen recognition. Thus, sensitive assays aimed at quantifying both replication-competent proviruses and defective, yet translationally competent proviruses are needed to understand the contribution of viral protein to HIV-1 pathogenesis and determine the effectiveness of HIV-1 cure interventions. Previously, we reported a modified HIV-1 gag p24 digital enzyme-linked immunosorbent assay with single molecule array (Simoa) detection of cell-associated viral protein. Here we report a novel p24 protein enrichment method coupled with the digital immunoassay to further extend the sensitivity and specificity of viral protein detection. Immunocapture of HIV gag p24 followed by elution in a Simoa-compatible format resulted in higher protein recovery and lower background from various biological matrices and sample volumes. Quantification of as little as 1 fg of p24 protein from cell lysates from cells isolated from peripheral blood or tissues from ART-suppressed HIV participants, as well as simian-human immunodeficiency virus-infected non-human primates (NHPs), with high recovery and reproducibility is demonstrated here. The application of these enhanced methods to patient-derived samples has potential to further the study of the persistent HIV state and examine in vitro response to therapies, as well as ex vivo study of translationally competent cells from a variety of donors.
Keywords: HIV; SIV; Simoa; biomarker; immunoprecipitation; p24; p27; rectal biopsy.
Copyright © 2021 Wu, Cheney, Huang, Hazuda, Howell and Zuck.
Conflict of interest statement
GW, CC, QH, DH, BH, and PZ were employed by Merck & Co., Inc.
Figures

Similar articles
-
Ultrasensitive Detection of p24 in Plasma Samples from People with Primary and Chronic HIV-1 Infection.J Virol. 2021 Jun 24;95(14):e0001621. doi: 10.1128/JVI.00016-21. Epub 2021 Jun 24. J Virol. 2021. PMID: 33952636 Free PMC article.
-
Measuring the Inducible, Replication-Competent HIV Reservoir Using an Ultra-Sensitive p24 Readout, the Digital ELISA Viral Outgrowth Assay.Front Immunol. 2020 Aug 6;11:1971. doi: 10.3389/fimmu.2020.01971. eCollection 2020. Front Immunol. 2020. PMID: 32849659 Free PMC article.
-
Ultrasensitive Immunoassay for Simian Immunodeficiency Virus p27CA.AIDS Res Hum Retroviruses. 2018 Nov;34(11):993-1001. doi: 10.1089/AID.2018.0075. Epub 2018 Jul 10. AIDS Res Hum Retroviruses. 2018. PMID: 29869527 Free PMC article.
-
More reliable diagnosis of infection with human immunodeficiency virus type 1 (HIV-1) by detection of antibody IgGs to pol and gag proteins of HIV-1 and p24 antigen of HIV-1 in urine, saliva, and/or serum with highly sensitive and specific enzyme immunoassay (immune complex transfer enzyme immunoassay): a review.J Clin Lab Anal. 1997;11(5):267-86. doi: 10.1002/(SICI)1098-2825(1997)11:5<267::AID-JCLA5>3.0.CO;2-4. J Clin Lab Anal. 1997. PMID: 9292394 Free PMC article. Review.
-
Measurement of HIV-1 p24 antigen by signal-amplification-boosted ELISA of heat-denatured plasma is a simple and inexpensive alternative to tests for viral RNA.AIDS Rev. 2002 Apr-Jun;4(2):83-92. AIDS Rev. 2002. PMID: 12152521 Review.
Cited by
-
The Role of Electrochemical Sensors in Enhancing HIV Detection.Curr HIV Res. 2025;23(1):2-13. doi: 10.2174/011570162X363311250206045837. Curr HIV Res. 2025. PMID: 39950463 Review.
-
Application of ultrasensitive digital ELISA for p24 enables improved evaluation of HIV-1 reservoir diversity and growth kinetics in viral outgrowth assays.Sci Rep. 2023 Jul 6;13(1):10958. doi: 10.1038/s41598-023-37223-9. Sci Rep. 2023. PMID: 37414788 Free PMC article.
-
Combined noncanonical NF-κB agonism and targeted BET bromodomain inhibition reverse HIV latency ex vivo.J Clin Invest. 2022 Apr 15;132(8):e157281. doi: 10.1172/JCI157281. J Clin Invest. 2022. PMID: 35426377 Free PMC article.
-
Single cell spatial profiling of FFPE splenic tissue from a humanized mouse model of HIV infection.Biomark Res. 2024 Oct 8;12(1):116. doi: 10.1186/s40364-024-00658-x. Biomark Res. 2024. PMID: 39380117 Free PMC article.
-
An Ultrasensitive p24 Assay to Measure HIV-1 in Diverse Biological Matrixes.Methods Mol Biol. 2024;2807:343-354. doi: 10.1007/978-1-0716-3862-0_23. Methods Mol Biol. 2024. PMID: 38743239
References
-
- Castagna A., Muccini C., Galli L., Bigoloni A., Poli A., Spagnuolo V., et al. (2019). Analytical treatment interruption in chronic HIV-1 infection: time and magnitude of viral rebound in adults with 10 years of undetectable viral load and low HIV-DNA (APACHE study). J. Antimicrob. Chemother. 74 2039–2046. 10.1093/jac/dkz138 - DOI - PubMed
-
- Chang L., Song L., Fournier D. R., Kan C. W., Patel P. P., Ferrell E. P., et al. (2013). Simple diffusion-constrained immunoassay for p24 protein with the sensitivity of nucleic acid amplification for detecting acute HIV infection. J. Virol. Methods. 188 153–160. 10.1016/j.jviromet.2012.08.017 - DOI - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

