Computer-aided diagnosis of masses in breast computed tomography imaging: deep learning model with combined handcrafted and convolutional radiomic features
- PMID: 33796604
- PMCID: PMC8005916
- DOI: 10.1117/1.JMI.8.2.024501
Computer-aided diagnosis of masses in breast computed tomography imaging: deep learning model with combined handcrafted and convolutional radiomic features
Abstract
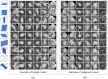
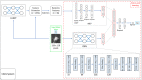
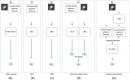
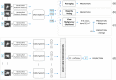
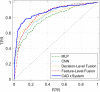
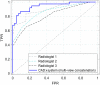
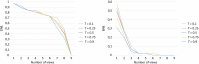
Purpose: A computer-aided diagnosis (CADx) system for breast masses is proposed, which incorporates both handcrafted and convolutional radiomic features embedded into a single deep learning model. Approach: The model combines handcrafted and convolutional radiomic signatures into a multi-view architecture, which retrieves three-dimensional (3D) image information by simultaneously processing multiple two-dimensional mass patches extracted along different planes through the 3D mass volume. Each patch is processed by a stream composed of two concatenated parallel branches: a multi-layer perceptron fed with automatically extracted handcrafted radiomic features, and a convolutional neural network, for which discriminant features are learned from the input patches. All streams are then concatenated together into a final architecture, where all network weights are shared and the learning occurs simultaneously for each stream and branch. The CADx system was developed and tested for diagnosis of breast masses ( ) using image datasets acquired with independent dedicated breast computed tomography systems from two different institutions. The diagnostic classification performance of the CADx system was compared against other machine and deep learning architectures adopting handcrafted and convolutional approaches, and three board-certified breast radiologists. Results: On a test set of 82 masses (45 benign, 37 malignant), the proposed CADx system performed better than all other model architectures evaluated, with an increase in the area under the receiver operating characteristics curve (AUC) of , and achieving a final AUC of 0.947, outperforming the three radiologists ( ). Conclusions: In conclusion, the system demonstrated its potential usefulness in breast cancer diagnosis by improving mass malignancy assessment.
Keywords: breast cancer; breast computed tomography; computer-aided diagnosis; deep learning; radiomics.
© 2021 Society of Photo-Optical Instrumentation Engineers (SPIE).
Figures


References
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

