Retention Using Selective Hooks (RUSH) Cargo Sorting Assay for Protein Vesicle Tracking in HeLa Cells
- PMID: 33796610
- PMCID: PMC8005885
- DOI: 10.21769/BioProtoc.3936
Retention Using Selective Hooks (RUSH) Cargo Sorting Assay for Protein Vesicle Tracking in HeLa Cells
Abstract
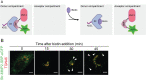
Monitoring vesicle trafficking is an excellent tool for the evaluation of protein dynamics in living cells. Such study is key for the understanding of protein sorting and secretion. Recent developments in microscopy, as well as new methodologies developed to study synchronized trafficking of proteins, allowed a better understanding of signaling, regulation and trafficking dynamics at the secretory pathway. One of the most helpful tools so far developed is the Retention Using Selective Hooks (RUSH) system, a methodology that facilitates the evaluation of synchronized cargo trafficking by monitoring fluorescent vesicles in cells upon biotin addition. Here we present a protocol that allows the quantitative evaluation of protein cargo trafficking at different fixed time points and an analytic approach that enables a better examination of specific cargo trafficking dynamics at the secretory pathway. Graphic abstract: Schematic representation of RUSH sorting assay in mammalian cells.
Keywords: Cargo sorting; Confocal microscopy; Protein trafficking; RUSH; Vesicle tracking.
Copyright © 2021 The Authors; exclusive licensee Bio-protocol LLC.
Conflict of interest statement
Competing interestsThe authors declare no competing financial interests.
Figures







References
-
- Boncompain G., Divoux S., Gareil N., de Forges H., Lescure A., Latreche L., Mercanti V., Jollivet F., Raposo G. and Perez F.(2012). Synchronization of secretory protein traffic in populations of cells. Nat Methods 9(5): 493-498. - PubMed
-
- Boncompain G. and Perez F.(2013). Fluorescence-based analysis of trafficking in mammalian cells. Methods Cell Biol 118: 179-194. - PubMed
-
- Boncompain G. and Weigel A. V.(2018). Transport and sorting in the Golgi complex: multiple mechanisms sort diverse cargo. Curr Opin Cell Biol 50: 94-101. - PubMed
-
- Crevenna A. H., Blank B., Maiser A., Emin D., Prescher J., Beck G., Kienzle C., Bartnik K., Habermann B., Pakdel M., Leonhardt H., Lamb D. C. and von Blume J.(2016). Secretory cargo sorting by Ca2+-dependent Cab45 oligomerization at the trans-Golgi network . J Cell Biol 213(3): 305-314. - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

