Exploring the landscape of ectodomain shedding by quantitative protein terminomics
- PMID: 33796845
- PMCID: PMC7995609
- DOI: 10.1016/j.isci.2021.102259
Exploring the landscape of ectodomain shedding by quantitative protein terminomics
Abstract
Ectodomain shedding is a proteolytic process that regulates the levels and functions of membrane proteins. Dysregulated shedding is linked to severe diseases, including cancer and Alzheimer's disease. However, the exact cleavage sites of shedding substrates remain largely unknown. Here, we explore the landscape of ectodomain shedding by generating large-scale, cell-type-specific maps of shedding cleavage sites. By means of N- and C-terminal peptide enrichment and quantitative mass spectrometry, we quantified protein termini in the culture media of 10 human cell lines and identified 489 cleavage sites on 163 membrane proteins whose proteolytic terminal fragments are downregulated in the presence of a broad-spectrum metalloprotease inhibitor. A major fraction of the presented cleavage sites was identified in a cell-type-specific manner and mapped onto receptors, cell adhesion molecules, and protein kinases and phosphatases. We confidently identified 86 cleavage sites as metalloprotease substrates by means of knowledge-based scoring.
Keywords: Cell Biology; Molecular Biology; Omics; Proteomics.
© 2021 The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



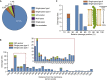
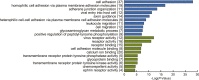



References
-
- Adachi J., Hashiguchi K., Nagano M., Sato M., Sato A., Fukamizu K., Ishihama Y., Tomonaga T. Improved proteome and phosphoproteome analysis on a cation exchanger by a combined acid and salt gradient. Anal. Chem. 2016;88:7899–7903. - PubMed
-
- Apweiler R., Hermjakob H., Sharon N. On the frequency of protein glycosylation, as deduced from analysis of the SWISS-PROT database. Biochim. Biophys. Acta. 1999;1473:4–8. - PubMed
-
- Beard H.A., Barniol-Xicota M., Yang J., Verhelst S.H.L. Discovery of cellular roles of intramembrane proteases. ACS Chem. Biol. 2019;14:2372–2388. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

