Linked transmission chains of imported SARS-CoV-2 variant B.1.351 across mainland France, January 2021
- PMID: 33797392
- PMCID: PMC8017907
- DOI: 10.2807/1560-7917.ES.2021.26.13.2100333
Linked transmission chains of imported SARS-CoV-2 variant B.1.351 across mainland France, January 2021
Abstract
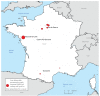
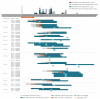
Two cases of confirmed SARS-CoV-2 infection with the B.1.351 variant were reported in France in mid-January, 2020. These cases attended a gathering in Mozambique in mid-December 2020. Investigations led to the identification of five imported cases responsible for 14 transmission chains and a total 36 cases. Epidemiological characteristics seemed comparable to those described before the emergence of the South African variant B.1.351. The lack of tertiary transmission outside of the personal sphere suggests that distancing and barrier measures were effective.
Keywords: COVID-19; Cluster; Epidemiology; Incubation; SARS-CoV-2 variants 20H/501Y.V2.
Conflict of interest statement
Figures
References
-
- O’Toole A, Scher E, Underwood A, Jackson B, Hill V, McCrone JT, et al. Pangolin: lineage assignment in an emerging pandemic as an epidemiological tool. github. [Accessed: 30 Mar2021]. Available from: https://cov-lineages.org/global_report_B.1.351.html - PMC - PubMed
-
- Bedford T, Hodcroft EB, Neher RA. Updated Nextstrain SARS-CoV-2 clade naming strategy. Nextstrain. [Accessed: 29 Mar 2021]. Available from: https://nextstrain.org/blog/2021-01-06-updated-SARS-CoV-2-clade-naming
-
- Tegally H, Wilkinson E, Giovanetti M, Iranzadeh A, Fonseca V, Giandhari J, et al. Emergence and rapid spread of a new severe acute respiratory syndrome-related coronavirus 2 (SARS-CoV-2) lineage with multiple spike mutations in South Africa. medRxiv 2020.12.21.20248640. https://doi.org/ 10.1101/2020.12.21.20248640 - DOI
-
- Vasques Nonaka CK, Miranda Franco M, Gräf T, Almeida Mendes AV, Santana de Aguiar R, Giovanetti M, et al. Genomic evidence of a Sars-Cov-2 reinfection case with E484K spike mutation in Brazil. Preprints. 2021;2021010132. 10.20944/preprints202101.0132.v1 - DOI
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous
