Epigenetic clock and methylation study of oocytes from a bovine model of reproductive aging
- PMID: 33797841
- PMCID: PMC8135012
- DOI: 10.1111/acel.13349
Epigenetic clock and methylation study of oocytes from a bovine model of reproductive aging
Abstract
Cattle are an attractive animal model of fertility in women due to their high degree of similarity relative to follicle selection, embryo cleavage, blastocyst formation, and gestation length. To facilitate future studies of the epigenetic underpinnings of aging effects in the female reproductive axis, several DNA methylation-based biomarkers of aging (epigenetic clocks) for bovine oocytes are presented. One such clock was germane to only oocytes, while a dual-tissue clock was highly predictive of age in both oocytes and blood. Dual species clocks that apply to both humans and cattle were also developed and evaluated. These epigenetic clocks can be used to accurately estimate the biological age of oocytes. Both epigenetic clock studies and epigenome-wide association studies revealed that blood and oocytes differ substantially with respect to aging and the underlying epigenetic signatures that potentially influence the aging process. The rate of epigenetic aging was found to be slower in oocytes compared to blood; however, oocytes appeared to begin at an older epigenetic age. The epigenetic clocks for oocytes are expected to address questions in the field of reproductive aging, including the central question: how to slow aging of oocytes.
Keywords: DNA methylation; epigenetic clock; epigenome-wide association study; oocytes; reproductive aging.
© 2021 The Authors. Aging Cell published by Anatomical Society and John Wiley & Sons Ltd.
Conflict of interest statement
SH is a founder of the non‐profit Epigenetic Clock Development Foundation which plans to license several patents from his employer UC Regents. These patents list SH as inventor. The other authors declare no conflicts of interest.
Figures
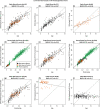
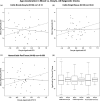



References
-
- Arneson, A. , Haghani, A. , Thompson, M. J. , Pellegrini, M. , Kwon, S. B. , Vu, H. , Li, C. Z. , Lu, A. T. , Barnes, B. , Hansen, K. D. , Zhou, W. , Breeze, C. E. , Ernst, J. , & Horvath, S. (2021). A mammalian methylation array for profiling methylation levels at conserved sequences. bioRxiv. https://www.biorxiv.org/content/10.1101/2021.01.07.425637v1 - DOI - PMC - PubMed
-
- Baired, D. T. , Collins, J. , Egozcue, J. , Evers, L. H. , Gianaroli, L. , Leridon, H. , Sunde, A. , Templeton, A. , Van Steirteghem, A. , Cohen, J. , Crosignani, P. G. , Devroey, P. , Diedrich, K. , Fauser, B. C. , Fraser, L. , Glasier, A. , Liebaers, I. , Mautone, G. , Penney, G. , & Tarlatzis, B. (2005). Fertility and ageing. Human Reproduction Update, 11(3), 261–276. 10.1093/humupd/dmi006 - DOI - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

