Discovering the landscape of protein modifications
- PMID: 33798408
- PMCID: PMC8106652
- DOI: 10.1016/j.molcel.2021.03.015
Discovering the landscape of protein modifications
Abstract
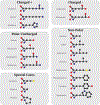
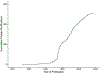
Protein modifications modulate nearly every aspect of cell biology in organisms, ranging from Archaea to Eukaryotes. The earliest evidence of covalent protein modifications was found in the early 20th century by studying the amino acid composition of proteins by chemical hydrolysis. These discoveries challenged what defined a canonical amino acid. The advent and rapid adoption of mass-spectrometry-based proteomics in the latter part of the 20th century enabled a veritable explosion in the number of known protein modifications, with more than 500 discrete modifications counted today. Now, new computational tools in data science, machine learning, and artificial intelligence are poised to allow researchers to make significant progress in discovering new protein modifications and determining their function. In this review, we take an opportunity to revisit the historical discovery of key post-translational modifications, quantify the current landscape of covalent protein adducts, and assess the role that new computational tools will play in the future of this field.
Keywords: amino acid; metabolism; post-translational modifications; protein acylation; protein modifications.
Copyright © 2021 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures



References
-
- Aebersold R, and Mann M (2003). Mass spectrometry-based proteomics. Nature 422, 198–207. - PubMed
-
- Alderson NL, Wang Y, Blatnik M, Frizzell N, Walla MD, Lyons TJ, Alt N, Carson JA, Nagai R, Thorpe SR, et al. (2006). S-(2-Succinyl)cysteine: a novel chemical modification of tissue proteins by a Krebs cycle intermediate. Arch Biochem Biophys 450, 1–8. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

