Opportunities and challenges for the computational interpretation of rare variation in clinically important genes
- PMID: 33798442
- PMCID: PMC8059338
- DOI: 10.1016/j.ajhg.2021.03.003
Opportunities and challenges for the computational interpretation of rare variation in clinically important genes
Abstract
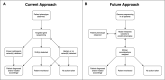
Genome sequencing is enabling precision medicine-tailoring treatment to the unique constellation of variants in an individual's genome. The impact of recurrent pathogenic variants is often understood, however there is a long tail of rare genetic variants that are uncharacterized. The problem of uncharacterized rare variation is especially acute when it occurs in genes of known clinical importance with functionally consequential variants and associated mechanisms. Variants of uncertain significance (VUSs) in these genes are discovered at a rate that outpaces current ability to classify them with databases of previous cases, experimental evaluation, and computational predictors. Clinicians are thus left without guidance about the significance of variants that may have actionable consequences. Computational prediction of the impact of rare genetic variation is increasingly becoming an important capability. In this paper, we review the technical and ethical challenges of interpreting the function of rare variants in two settings: inborn errors of metabolism in newborns and pharmacogenomics. We propose a framework for a genomic learning healthcare system with an initial focus on early-onset treatable disease in newborns and actionable pharmacogenomics. We argue that (1) a genomic learning healthcare system must allow for continuous collection and assessment of rare variants, (2) emerging machine learning methods will enable algorithms to predict the clinical impact of rare variants on protein function, and (3) ethical considerations must inform the construction and deployment of all rare-variation triage strategies, particularly with respect to health disparities arising from unbalanced ancestry representation.
Copyright © 2021 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
The authors declare no competing interests.
Figures


References
-
- Richards S., Aziz N., Bale S., Bick D., Das S., Gastier-Foster J., Grody W.W., Hegde M., Lyon E., Spector E., ACMG Laboratory Quality Assurance Committee Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015;17:405–424. - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

