The Time Has Come to Explore Plasma Biomarkers in Genetic Cardiomyopathies
- PMID: 33799487
- PMCID: PMC7998409
- DOI: 10.3390/ijms22062955
The Time Has Come to Explore Plasma Biomarkers in Genetic Cardiomyopathies
Abstract
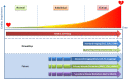
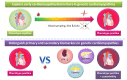
For patients with hypertrophic cardiomyopathy (HCM), dilated cardiomyopathy (DCM) or arrhythmogenic cardiomyopathy (ACM), screening for pathogenic variants has become standard clinical practice. Genetic cascade screening also allows the identification of relatives that carry the same mutation as the proband, but disease onset and severity in mutation carriers often remains uncertain. Early detection of disease onset may allow timely treatment before irreversible changes are present. Although plasma biomarkers may aid in the prediction of disease onset, monitoring relies predominantly on identifying early clinical symptoms, on imaging techniques like echocardiography (Echo) and cardiac magnetic resonance imaging (CMR), and on (ambulatory) electrocardiography (electrocardiograms (ECGs)). In contrast to most other cardiac diseases, which are explained by a combination of risk factors and comorbidities, genetic cardiomyopathies have a clear primary genetically defined cardiac background. Cardiomyopathy cohorts could therefore have excellent value in biomarker studies and in distinguishing biomarkers related to the primary cardiac disease from those related to extracardiac, secondary organ dysfunction. Despite this advantage, biomarker investigations in cardiomyopathies are still limited, most likely due to the limited number of carriers in the past. Here, we discuss not only the potential use of established plasma biomarkers, including natriuretic peptides and troponins, but also the use of novel biomarkers, such as cardiac autoantibodies in genetic cardiomyopathy, and discuss how we can gauge biomarker studies in cardiomyopathy cohorts for heart failure at large.
Keywords: ACM; DCM; HCM; cardiac autoantibodies; early detection; genetic cardiomyopathy; noncoding RNA; plasma biomarkers.
Conflict of interest statement
The UMCG, which employs the authors, has received research grants and/or fees from AstraZeneca, Abbott, Bristol-Myers Squibb, Novartis, Novo Nordisk and Roche. de Boer received speaker fees from Abbott, AstraZeneca, Bayer, Novartis and Roche.
Figures


References
-
- Richardson P., McKenna R.W., Bristow M., Maisch B., Mautner B., O’Connell J., Olsen E., Thiene G., Goodwin J., Gyarfas I., et al. Report of the 1995 World Health Organization/International Society and Federation of Cardiology Task Force on the Definition and Classification of Cardiomyopathies. Circulation. 1996;93:841–842. doi: 10.1161/01.CIR.93.5.841. - DOI - PubMed
-
- Pinto Y.M., Elliott P.M., Arbustini E., Adler Y., Anastasakis A., Böhm M., Duboc D., Gimeno J., de Groote P., Imazio M., et al. Proposal for a revised definition of dilated cardiomyopathy, hypokinetic non-dilated cardiomyopathy, and its implications for clinical practice: A position statement of the ESC working group on myocardial and pericardial diseases. Eur. Heart J. 2016;37:1850–1858. doi: 10.1093/eurheartj/ehv727. - DOI - PubMed
-
- Mestroni L., Rocco C., Gregori D., Sinagra G., Di Lenarda A., Miocic S., Vatta M., Pinamonti B., Muntoni F., Caforio A.L., et al. Familial dilated cardiomyopathy: Evidence for genetic and phenotypic heterogeneity. Heart Muscle Disease Study Group. J. Am. Coll. Cardiol. 1999;34:181–190. doi: 10.1016/S0735-1097(99)00172-2. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

