In Silico Analysis of P450s and Their Role in Secondary Metabolism in the Bacterial Class Gammaproteobacteria
- PMID: 33799696
- PMCID: PMC7998510
- DOI: 10.3390/molecules26061538
In Silico Analysis of P450s and Their Role in Secondary Metabolism in the Bacterial Class Gammaproteobacteria
Abstract
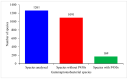
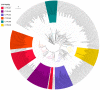
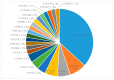
The impact of lifestyle on shaping the genome content of an organism is a well-known phenomenon and cytochrome P450 enzymes (CYPs/P450s), heme-thiolate proteins that are ubiquitously present in organisms, are no exception. Recent studies focusing on a few bacterial species such as Streptomyces, Mycobacterium, Cyanobacteria and Firmicutes revealed that the impact of lifestyle affected the P450 repertoire in these species. However, this phenomenon needs to be understood in other bacterial species. We therefore performed genome data mining, annotation, phylogenetic analysis of P450s and their role in secondary metabolism in the bacterial class Gammaproteobacteria. Genome-wide data mining for P450s in 1261 Gammaproteobacterial species belonging to 161 genera revealed that only 169 species belonging to 41 genera have P450s. A total of 277 P450s found in 169 species grouped into 84 P450 families and 105 P450 subfamilies, where 38 new P450 families were found. Only 18% of P450s were found to be involved in secondary metabolism in Gammaproteobacterial species, as observed in Firmicutes as well. The pathogenic or commensal lifestyle of Gammaproteobacterial species influences them to such an extent that they have the lowest number of P450s compared to other bacterial species, indicating the impact of lifestyle on shaping the P450 repertoire. This study is the first report on comprehensive analysis of P450s in Gammaproteobacteria.
Keywords: Cyanobacteria; Firmicutes; Gammaproteobacteria; Mycobacterium; Streptomyces; biosynthetic gene clusters; cytochrome P450 monooxygenases; genome data mining; lifestyle; secondary metabolism.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures

References
-
- Berman J.J. Taxonomic Guide to Infectious Diseases: Understanding the Biologic Classes of Pathogenic Organisms. Academic Press; Cambridge, MA, USA: 2019.
-
- Kersters K., De Vos P., Gillis M., Swings J., Vandamme P., Stackebrandt E. The Prokaryotes: A Handbook on the Biology of Bacteria. Volume 5. Springer; New York, NY, USA: 2006. Introduction to the Proteobacteria; pp. 3–37.
-
- Williams K.P., Kelly D.P. Proposal for a new class within the phylum Proteobacteria, Acidithiobacillia classis nov., with the type order Acidithiobacillales, and emended description of the class Gammaproteobacteria. Int. J. Syst. Evol. Microbiol. 2013;63:2901–2906. doi: 10.1099/ijs.0.049270-0. - DOI - PubMed
-
- Liu L., Chen D., Liu L., Lan R., Hao S., Jin W., Sun H., Wang Y., Liang Y., Xu J. Genetic diversity, multidrug resistance, and virulence of Citrobacter freundii from diarrheal patients and healthy individuals. Front. Cell. Infect. Microbiol. 2018;8:233. doi: 10.3389/fcimb.2018.00233. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

