A Survey of Transposon Landscapes in the Putative Ancient Asexual Ostracod Darwinula stevensoni
- PMID: 33799706
- PMCID: PMC7998251
- DOI: 10.3390/genes12030401
A Survey of Transposon Landscapes in the Putative Ancient Asexual Ostracod Darwinula stevensoni
Abstract
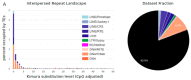
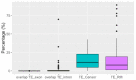
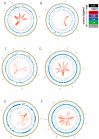
How asexual reproduction shapes transposable element (TE) content and diversity in eukaryotic genomes remains debated. We performed an initial survey of TE load and diversity in the putative ancient asexual ostracod Darwinula stevensoni. We examined long contiguous stretches of DNA in clones from a genomic fosmid library, totaling about 2.5 Mb, and supplemented these data with results on TE abundance and diversity from an Illumina draft genome. In contrast to other TE studies in putatively ancient asexuals, which revealed relatively low TE content, we found that at least 19% of the fosmid dataset and 26% of the genome assembly corresponded to known transposons. We observed a high diversity of transposon families, including LINE, gypsy, PLE, mariner/Tc, hAT, CMC, Sola2, Ginger, Merlin, Harbinger, MITEs and helitrons, with the prevalence of DNA transposons. The predominantly low levels of sequence diversity indicate that many TEs are or have recently been active. In the fosmid data, no correlation was found between telomeric repeats and non-LTR retrotransposons, which are present near telomeres in other taxa. Most TEs in the fosmid data were located outside of introns and almost none were found in exons. We also report an N-terminal Myb/SANT-like DNA-binding domain in site-specific R4/Dong non-LTR retrotransposons. Although initial results on transposable loads need to be verified with high quality draft genomes, this study provides important first insights into TE dynamics in putative ancient asexual ostracods.
Keywords: DNA transposons; asexuality; crustaceans; retrotransposons; transposable elements.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

