The Landscape of Signaling Pathways and Proteasome Inhibitors Combinations in Multiple Myeloma
- PMID: 33799793
- PMCID: PMC8000754
- DOI: 10.3390/cancers13061235
The Landscape of Signaling Pathways and Proteasome Inhibitors Combinations in Multiple Myeloma
Abstract
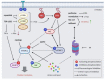
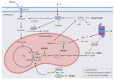
Multiple myeloma is a malignancy of terminally differentiated plasma cells, characterized by an extreme genetic heterogeneity that poses great challenges for its successful treatment. Due to antibody overproduction, MM cells depend on the precise regulation of the protein degradation systems. Despite the success of PIs in MM treatment, resistance and adverse toxic effects such as peripheral neuropathy and cardiotoxicity could arise. To this end, the use of rational combinatorial treatments might allow lowering the dose of inhibitors and therefore, minimize their side-effects. Even though the suppression of different cellular pathways in combination with proteasome inhibitors have shown remarkable anti-myeloma activities in preclinical models, many of these promising combinations often failed in clinical trials. Substantial progress has been made by the simultaneous targeting of proteasome and different aspects of MM-associated immune dysfunctions. Moreover, targeting deranged metabolic hubs could represent a new avenue to identify effective therapeutic combinations with PIs. Finally, epigenetic drugs targeting either DNA methylation, histone modifiers/readers, or chromatin remodelers are showing pleiotropic anti-myeloma effects alone and in combination with PIs. We envisage that the positive outcome of patients will probably depend on the availability of more effective drug combinations and treatment of early MM stages. Therefore, the identification of sensitive targets and aberrant signaling pathways is instrumental for the development of new personalized therapies for MM patients.
Keywords: combinatorial treatment; drug resistance; multiple myeloma; proteasome inhibitors; synthetic lethality.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Kumar S., Paiva B., Anderson K.C., Durie B., Landgren O., Moreau P., Munshi N., Lonial S., Bladé J., Mateos M.-V., et al. International Myeloma Working Group consensus criteria for response and minimal residual disease assessment in multiple myeloma. Lancet Oncol. 2016;17:e328–e346. doi: 10.1016/s1470-2045(16)30206-6. - DOI - PubMed
-
- Maura F., Bolli N., Angelopoulos N., Dawson K.J., Leongamornlert D., Martincorena I., Mitchell T.J., Fullam A., Gonzalez S., Szalat R., et al. Genomic landscape and chronological reconstruction of driver events in multiple myeloma. Nat. Commun. 2019;10:1–12. doi: 10.1038/s41467-019-11680-1. - DOI - PMC - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

