PARP7 and Mono-ADP-Ribosylation Negatively Regulate Estrogen Receptor α Signaling in Human Breast Cancer Cells
- PMID: 33799807
- PMCID: PMC8001409
- DOI: 10.3390/cells10030623
PARP7 and Mono-ADP-Ribosylation Negatively Regulate Estrogen Receptor α Signaling in Human Breast Cancer Cells
Abstract
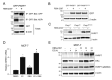
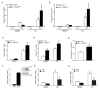
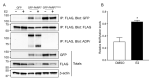
ADP-ribosylation is a post-translational protein modification catalyzed by a family of proteins known as poly-ADP-ribose polymerases. PARP7 (TIPARP; ARTD14) is a mono-ADP-ribosyltransferase involved in several cellular processes, including responses to hypoxia, innate immunity and regulation of nuclear receptors. Since previous studies suggested that PARP7 was regulated by 17β-estradiol, we investigated whether PARP7 regulates estrogen receptor α signaling. We confirmed the 17β-estradiol-dependent increases of PARP7 mRNA and protein levels in MCF-7 cells, and observed recruitment of estrogen receptor α to the promoter of PARP7. Overexpression of PARP7 decreased ligand-dependent estrogen receptor α signaling, while treatment of PARP7 knockout MCF-7 cells with 17β-estradiol resulted in increased expression of and recruitment to estrogen receptor α target genes, in addition to increased proliferation. Co-immunoprecipitation assays revealed that PARP7 mono-ADP-ribosylated estrogen receptor α, and mass spectrometry mapped the modified peptides to the receptor's ligand-independent transactivation domain. Co-immunoprecipitation with truncated estrogen receptor α variants identified that the hinge region of the receptor is required for PARP7-dependent mono-ADP-ribosylation. These results imply that PARP7-mediated mono-ADP-ribosylation may play an important role in estrogen receptor positive breast cancer.
Keywords: ARTD14; PARP7; TIPARP; breast cancer; estrogen receptor α; mono-ADP-ribosylation; poly ADP-ribose polymerase.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

