The Role of Autophagy and lncRNAs in the Maintenance of Cancer Stem Cells
- PMID: 33799834
- PMCID: PMC7998932
- DOI: 10.3390/cancers13061239
The Role of Autophagy and lncRNAs in the Maintenance of Cancer Stem Cells
Abstract
Cancer stem cells (CSCs) possess properties such as self-renewal, resistance to apoptotic cues, quiescence, and DNA-damage repair capacity. Moreover, CSCs strongly influence the tumour microenvironment (TME) and may account for cancer progression, recurrence, and relapse. CSCs represent a distinct subpopulation in tumours and the detection, characterisation, and understanding of the regulatory landscape and cellular processes that govern their maintenance may pave the way to improving prognosis, selective targeted therapy, and therapy outcomes. In this review, we have discussed the characteristics of CSCs identified in various cancer types and the role of autophagy and long noncoding RNAs (lncRNAs) in maintaining the homeostasis of CSCs. Further, we have discussed methods to detect CSCs and strategies for treatment and relapse, taking into account the requirement to inhibit CSC growth and survival within the complex backdrop of cellular processes, microenvironmental interactions, and regulatory networks associated with cancer. Finally, we critique the computationally reinforced triangle of factors inclusive of CSC properties, the process of autophagy, and lncRNA and their associated networks with respect to hypoxia, epithelial-to-mesenchymal transition (EMT), and signalling pathways.
Keywords: LncRNAs; autophagy; cancer stem cells (CSCs); haematological malignancies; solid cancers; tumour microenvironment.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


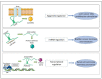

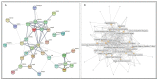
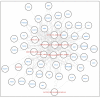
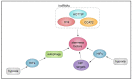
References
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources

