Structure-Based Virtual Screening Identifies Multiple Stable Binding Sites at the RecA Domains of SARS-CoV-2 Helicase Enzyme
- PMID: 33800013
- PMCID: PMC7962107
- DOI: 10.3390/molecules26051446
Structure-Based Virtual Screening Identifies Multiple Stable Binding Sites at the RecA Domains of SARS-CoV-2 Helicase Enzyme
Abstract
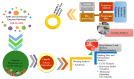
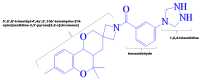
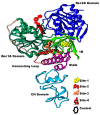
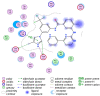
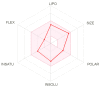
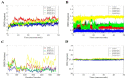
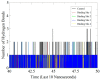
With the emergence and global spread of the COVID-19 pandemic, the scientific community worldwide has focused on search for new therapeutic strategies against this disease. One such critical approach is targeting proteins such as helicases that regulate most of the SARS-CoV-2 RNA metabolism. The purpose of the current study was to predict a library of phytochemicals derived from diverse plant families with high binding affinity to SARS-CoV-2 helicase (Nsp13) enzyme. High throughput virtual screening of the Medicinal Plant Database for Drug Design (MPD3) database was performed on SARS-CoV-2 helicase using AutoDock Vina. Nilotinib, with a docking value of -9.6 kcal/mol, was chosen as a reference molecule. A compound (PubChem CID: 110143421, ZINC database ID: ZINC257223845, eMolecules: 43290531) was screened as the best binder (binding energy of -10.2 kcal/mol on average) to the enzyme by using repeated docking runs in the screening process. On inspection, the compound was disclosed to show different binding sites of the triangular pockets collectively formed by Rec1A, Rec2A, and 1B domains and a stalk domain at the base. The molecule is often bound to the ATP binding site (referred to as binding site 2) of the helicase enzyme. The compound was further discovered to fulfill drug-likeness and lead-likeness criteria, have good physicochemical and pharmacokinetics properties, and to be non-toxic. Molecular dynamic simulation analysis of the control/lead compound complexes demonstrated the formation of stable complexes with good intermolecular binding affinity. Lastly, affirmation of the docking simulation studies was accomplished by estimating the binding free energy by MMPB/GBSA technique. Taken together, these findings present further in silco investigation of plant-derived lead compounds to effectively address COVID-19.
Keywords: COVID-19; SARS-CoV-2 helicase; molecular dynamic simulation; phytochemicals.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

