Identification of Enterotype and Its Effects on Intestinal Butyrate Production in Pigs
- PMID: 33800148
- PMCID: PMC7999521
- DOI: 10.3390/ani11030730
Identification of Enterotype and Its Effects on Intestinal Butyrate Production in Pigs
Abstract
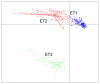
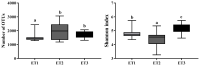
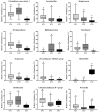
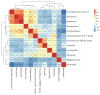
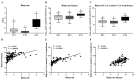
Gut microbiota is thought to play a crucial role in nutrient digestion for pigs, especially in processing indigestible polysaccharides in the diets to produce short-chain fatty acids (SCFAs). However, the link between microbiota community structure and phenotypic performances are poorly understood. In the present study, the fecal samples of 105 Jinhua pigs at 105 days of age were clustered into three enterotypes (ETs, ET1, ET2, and ET3) that are subpopulations of distinct bacterial community composition by using 16S rRNA high throughput sequencing. The α-diversity indices (the OTU number and Shannon index) were significantly different among the ETs (p < 0.001). At the genus level, the ET1 group was over-represented by Lactobacillus (17.49%) and Clostridium sensu stricto 1 (11.78%), the ET2 group was over-represented by Clostridium sensu stricto 1 (17.49%) and Bifidobacterium (11.78%), and the ET3 group was over-represented by Bacteroides (18.17%). Significant differences in the fecal contents of butyrate were observed among ETs, with the highest level detected in ET3 and the lowest in ET2 (p < 0.05). Consistently, more copies of the terminal genes for butyrate synthesis, butyrate kinase (Buk) and butyryl coenzyme A (CoA): acetate CoA transferase (But) were detected by qPCR in the fecal samples of the ET3 group as compared to other two groups (p < 0.05). In addition, of the two genes, But was demonstrated to be more relevant to the butyrate content (R = 0.7464) than Buk (R = 0.4905) by correlation analysis. In addition, based on the taxonomic analysis, we found that Faecalibacterium was the most relevant butyrate-producing genera with fecal butyrate contents in Jinhua pigs, followed by Butyricicoccus, Eubacterium, Butyricimonas, Blautia, and Anaerostipes, all of which showed significantly higher richness in ET3 than as compared to ET1 and ET2 (p < 0.05). Collectively, this work presents a first overview of the enterotypes clustering in Jinhua pigs and will help to unravel the functional implications of ETs for the pig's phenotypic performance and nutrient metabolism.
Keywords: Jinhua pig; butyrate; enterotype; gut microbiota.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Enterotype identification and its influence on regulating the duodenum metabolism in chickens.Poult Sci. 2020 Mar;99(3):1515-1527. doi: 10.1016/j.psj.2019.10.078. Epub 2020 Jan 30. Poult Sci. 2020. PMID: 32111319 Free PMC article.
-
Core gut microbiota in Jinhua pigs and its correlation with strain, farm and weaning age.J Microbiol. 2018 May;56(5):346-355. doi: 10.1007/s12275-018-7486-8. Epub 2018 May 2. J Microbiol. 2018. PMID: 29721832
-
Effect of chronic and acute heat challenges on fecal microbiota composition, production, and thermoregulation traits in growing pigs1,2.J Anim Sci. 2019 Sep 3;97(9):3845-3858. doi: 10.1093/jas/skz222. J Anim Sci. 2019. PMID: 31268142 Free PMC article.
-
Butyrate producers, "The Sentinel of Gut": Their intestinal significance with and beyond butyrate, and prospective use as microbial therapeutics.Front Microbiol. 2023 Jan 12;13:1103836. doi: 10.3389/fmicb.2022.1103836. eCollection 2022. Front Microbiol. 2023. PMID: 36713166 Free PMC article. Review.
-
Bridging the Gap from Enterotypes to Personalized Dietary Recommendations: A Metabolomics Perspective on Microbiome Research.Metabolites. 2023 Dec 2;13(12):1182. doi: 10.3390/metabo13121182. Metabolites. 2023. PMID: 38132864 Free PMC article. Review.
Cited by
-
Rumen bacterial cluster identification and its influence on rumen metabolites and growth performance of young goats.Anim Nutr. 2023 Jul 20;15:34-44. doi: 10.1016/j.aninu.2023.05.013. eCollection 2023 Dec. Anim Nutr. 2023. PMID: 37771855 Free PMC article.
-
High-tannin food enhances spatial memory and scatter-hoarding in rodents via the microbiota-gut-brain axis.Microbiome. 2024 Jul 29;12(1):140. doi: 10.1186/s40168-024-01849-2. Microbiome. 2024. PMID: 39075602 Free PMC article.
-
Microbial signatures and enterotype clusters in fattening pigs: implications for nitrogen utilization efficiency.Front Microbiol. 2024 Apr 10;15:1354537. doi: 10.3389/fmicb.2024.1354537. eCollection 2024. Front Microbiol. 2024. PMID: 38659980 Free PMC article.
-
A first characterization of the microbiota-resilience link in swine.Microbiome. 2024 Mar 15;12(1):53. doi: 10.1186/s40168-024-01771-7. Microbiome. 2024. PMID: 38486255 Free PMC article.
-
Uncovering the biogeography of the microbial commmunity and its association with nutrient metabolism in the intestinal tract using a pig model.Front Nutr. 2022 Sep 26;9:1003763. doi: 10.3389/fnut.2022.1003763. eCollection 2022. Front Nutr. 2022. PMID: 36238459 Free PMC article.
References
Grants and funding
- 2010DS700124-ZZ1905/national nature science foundation of china : 31972999 State Key Laboratory for Managing Biotic and Chemical Threats to the Quality and Safety of Agro-products
- 2018KLGM04/Open project of key laboratory of Intestinal microecology in Zhejiang Province
- [2018]2279/Science and Technology Support Prject of GuiZhou province
LinkOut - more resources
Full Text Sources
Other Literature Sources

