Identification of Overexpressed Genes in Malignant Pleural Mesothelioma
- PMID: 33800494
- PMCID: PMC7962966
- DOI: 10.3390/ijms22052738
Identification of Overexpressed Genes in Malignant Pleural Mesothelioma
Abstract
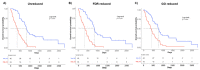
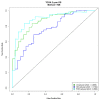
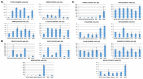
Malignant pleural mesothelioma (MPM) is a fatal tumor lacking effective therapies. The characterization of overexpressed genes could constitute a strategy for identifying drivers of tumor progression as targets for novel therapies. Thus, we performed an integrated gene-expression analysis on RNAseq data of 85 MPM patients from TCGA dataset and reference samples from the GEO. The gene list was further refined by using published studies, a functional enrichment analysis, and the correlation between expression and patients' overall survival. Three molecular signatures defined by 15 genes were detected. Seven genes were involved in cell adhesion and extracellular matrix organization, with the others in control of the mitotic cell division or apoptosis inhibition. Using Western blot analyses, we found that ADAMTS1, PODXL, CIT, KIF23, MAD2L1, TNNT1, and TRAF2 were overexpressed in a limited number of cell lines. On the other hand, interestingly, CTHRC1, E-selectin, SPARC, UHRF1, PRSS23, BAG2, and MDK were abundantly expressed in over 50% of the six MPM cell lines analyzed. Thus, these proteins are candidates as drivers for sustaining the tumorigenic process. More studies with small-molecule inhibitors or silencing RNAs are fully justified and need to be undertaken to better evaluate the cancer-driving role of the targets herewith identified.
Keywords: MPM; RNAseq; gene signature; malignant pleural mesothelioma; overexpressed genes; therapeutic targets.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
-
- McCambridge A.J., Napolitano A., Mansfield A.S., Fennell D.A., Sekido Y., Nowak A.K., Reungwetwattana T., Mao W., Pass H.I., Carbone M., et al. Progress in the Management of Malignant Pleural Mesothelioma in 2017. J. Thorac. Oncol. 2018;13:606–623. doi: 10.1016/j.jtho.2018.02.021. - DOI - PMC - PubMed
-
- Vogelzang N.J., Rusthoven J.J., Symanowski J., Denham C., Kaukel E., Ruffie P., Gatzemeier U., Boyer M., Emri S., Manegold C., et al. Phase III Study of Pemetrexed in Combination with Cisplatin Versus Cisplatin Alone in Patients with Malignant Pleural Mesothelioma. J. Clin. Oncol. 2003;21:2636–2644. doi: 10.1200/JCO.2003.11.136. - DOI - PubMed
-
- Hmeljak J., Sanchez-Vega F., Hoadley K.A., Shih J., Stewart C., Heiman D., Tarpey P., Danilova L., Drill E., Gibb E.A., et al. Integrative Molecular Characterization of Malignant Pleural Mesothelioma. Cancer Discov. 2018;8:1548–1565. doi: 10.1158/2159-8290.CD-18-0804. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

