Recent Progress in Torovirus Molecular Biology
- PMID: 33800523
- PMCID: PMC7998386
- DOI: 10.3390/v13030435
Recent Progress in Torovirus Molecular Biology
Abstract
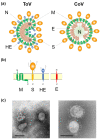
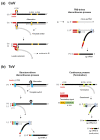
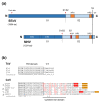
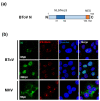
Torovirus (ToV) has recently been classified into the new family Tobaniviridae, although it belonged to the Coronavirus (CoV) family historically. ToVs are associated with enteric diseases in animals and humans. In contrast to CoVs, which are recognised as pathogens of veterinary and medical importance, little attention has been paid to ToVs because their infections are usually asymptomatic or not severe; for a long time, only one equine ToV could be propagated in cultured cells. However, bovine ToVs, which predominantly cause diarrhoea in calves, have been detected worldwide, leading to economic losses. Porcine ToVs have also spread globally; although they have not caused serious economic losses, coinfections with other pathogens can exacerbate their symptoms. In addition, frequent inter- or intra-recombination among ToVs can increase pathogenesis or unpredicted host adaptation. These findings have highlighted the importance of ToVs as pathogens and the need for basic ToV research. Here, we review recent progress in the study of ToV molecular biology including reverse genetics, focusing on the similarities and differences between ToVs and CoVs.
Keywords: coronavirus; enteric diseases; non-structural proteins; replication; reverse genetics; structural proteins; torovirus; transcription.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- ICTV (International Committee on Taxonomy of Virus) Virus Taxonomy: 2019 Release. ICTV; 2019.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

