Taxonomic Identification of Different Species of the Genus Aeromonas by Whole-Genome Sequencing and Use of Their Species-Specific β-Lactamases as Phylogenetic Markers
- PMID: 33800590
- PMCID: PMC8065696
- DOI: 10.3390/antibiotics10040354
Taxonomic Identification of Different Species of the Genus Aeromonas by Whole-Genome Sequencing and Use of Their Species-Specific β-Lactamases as Phylogenetic Markers
Abstract
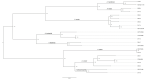
Some Aeromonas species, potentially pathogenic for humans, are known to express up to three different classes of chromosomal β-lactamases, which may become hyperproduced and cause treatment failure. The aim of this study was to assess the utility of these species-specific β-lactamase genes as phylogenetic markers using whole-genome sequencing data. Core-genome alignments were generated for 36 Aeromonas genomes from seven different species and scanned for antimicrobial resistance genes. Core-genome alignment confirmed the MALDI-TOF identification of most of the isolates and re-identified an A. hydrophila isolate as A. dhakensis. Three (B, C and D) of the four Ambler classes of β-lactamase genes were found in A. sobria, A. allosacharophila, A. hydrophila and A. dhakensis (blaCphA, blaAmpC and blaOXA). A. veronii only showed class-B- and class-D-like matches (blaCphA and blaOXA), whereas those for A. media, A. rivipollensis and A. caviae were class C and D (blaCMY, blaMOX and blaOXA427). The phylogenetic tree derived from concatenated sequences of β-lactamase genes successfully clustered each species. Some isolates also had resistance to sulfonamides, quinolones and aminoglycosides. Whole-genome sequencing proved to be a useful method to identify Aeromonas at the species level, which led to the unexpected identification of A. dhakensis and A.rivipollensis and revealed the resistome of each isolate.
Keywords: Aeromonas dhakensis; Aeromonas rivipollensis; FOX; MOX; beta-lactamases; cephamicinases; core-genome.
Conflict of interest statement
The authors declare no competing financial interest.
Figures
References
-
- Martin-Carnahan A., Joseph S.W. Order XII. Aeromonadales ord. nov. In: Brenner D.J., Krieg N.R., Staley J.T., Garrity G.M., editors. Bergey’s Manual of Systematic Bacteriology. 2nd ed. Williams & Wilkins; Philadelphia, PA, USA: 2005. pp. 556–578.
LinkOut - more resources
Full Text Sources
Other Literature Sources

