Na+/K+-ATPase Revisited: On Its Mechanism of Action, Role in Cancer, and Activity Modulation
- PMID: 33800655
- PMCID: PMC8061769
- DOI: 10.3390/molecules26071905
Na+/K+-ATPase Revisited: On Its Mechanism of Action, Role in Cancer, and Activity Modulation
Abstract
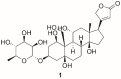
Maintenance of Na+ and K+ gradients across the cell plasma membrane is an essential process for mammalian cell survival. An enzyme responsible for this process, sodium-potassium ATPase (NKA), has been currently extensively studied as a potential anticancer target, especially in lung cancer and glioblastoma. To date, many NKA inhibitors, mainly of natural origin from the family of cardiac steroids (CSs), have been reported and extensively studied. Interestingly, upon CS binding to NKA at nontoxic doses, the role of NKA as a receptor is activated and intracellular signaling is triggered, upon which cancer cell death occurs, which lies in the expression of different NKA isoforms than in healthy cells. Two major CSs, digoxin and digitoxin, originally used for the treatment of cardiac arrhythmias, are also being tested for another indication-cancer. Such drug repositioning has a big advantage in smoother approval processes. Besides this, novel CS derivatives with improved performance are being developed and evaluated in combination therapy. This article deals with the NKA structure, mechanism of action, activity modulation, and its most important inhibitors, some of which could serve not only as a powerful tool to combat cancer, but also help to decipher the so-far poorly understood NKA regulation.
Keywords: Na+/K+-ATPase activity modulation; anticancer activity; cardiac glycosides; combination therapy; digitoxigenin; digitoxin; digoxin; natural compounds; ouabain; sodium-potassium pump inhibitors.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
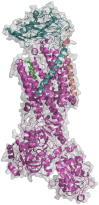


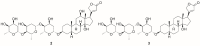
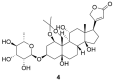
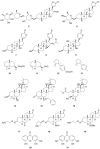
References
-
- FDA. [(accessed on 13 October 2020)]; Available online: https://www.fda.gov/news-events/press-announcements/fda-approves-first-p....
-
- McCarthy M.J., Pagba C.V., Prakash P., Naji A.K., van der Hoeven D., Liang H., Gupta A.K., Zhou Y., Cho K.J., Hancock J.F. Discovery of high-affinity noncovalent allosteric KRAS inhibitors that disrupt effector binding. ACS Omega. 2019;4:2921–2930. doi: 10.1021/acsomega.8b03308. - DOI - PMC - PubMed
-
- Guimarães I.S., Daltoé R.D., Herlinger A.L., Madeira K.P., Ladislau T., Valadão J.C., Lyra P.C.M., Jr., Teixeira S.F., Amorim G.M., dos Santos D.Z., et al. Conventional cancer treatment. In: Rangel L., editor. Cancer Treatment—Conventional and Innovative Approaches. IntechOpen; London, UK: 2013. pp. 3–35. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

