A Genomic Blueprint of Flax Fungal Parasite Fusarium oxysporum f. sp. lini
- PMID: 33800857
- PMCID: PMC7961770
- DOI: 10.3390/ijms22052665
A Genomic Blueprint of Flax Fungal Parasite Fusarium oxysporum f. sp. lini
Abstract
Fusarium wilt of flax is an aggressive disease caused by the soil-borne fungal pathogen Fusarium oxysporum f. sp. lini. It is a challenging pathogen presenting a constant threat to flax production industry worldwide. Previously, we reported chromosome-level assemblies of 5 highly pathogenic F. oxysporum f. sp. lini strains. We sought to characterize the genomic architecture of the fungus and outline evolutionary mechanisms shaping the pathogen genome. Here, we reveal the complex multi-compartmentalized genome organization and uncover its diverse evolutionary dynamics, which boosts genetic diversity and facilitates host adaptation. In addition, our results suggest that host of functions implicated in the life cycle of mobile genetic elements are main contributors to dissimilarity between proteomes of different Fusaria. Finally, our experiments demonstrate that mobile genetics elements are expressed in planta upon infection, alluding to their role in pathogenicity. On the whole, these results pave the way for further in-depth studies of evolutionary forces shaping the host-pathogen interaction.
Keywords: Fusarium oxysporum f. sp. lini; comparative genomics; flax; genome architecture; genome evolution; transposable elements.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
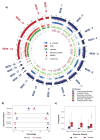


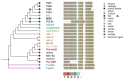
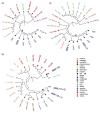

References
-
- Zhang Y., Ma L.-J. Deciphering Pathogenicity of Fusarium oxysporum From a Phylogenomics Perspective. Adv. Genet. 2017;100:179–209. - PubMed
-
- Williams A.H., Sharma M., Thatcher L.F., Azam S., Hane J.K., Sperschneider J., Kidd B.N., Anderson J.P., Ghosh R., Garg G., et al. Comparative genomics and prediction of conditionally dispensable sequences in legume–infecting Fusarium oxysporum formae speciales facilitates identification of candidate effectors. BMC Genom. 2016;17:311–324. doi: 10.1186/s12864-016-2486-8. - DOI - PMC - PubMed
-
- Armitage A.D., Taylor A., Sobczyk M.K., Baxter L., Greenfield B.P.J., Bates H.J., Wilson F., Jackson A.C., Ott S., Harrison R.J., et al. Characterisation of pathogen-specific regions and novel effector candidates in Fusarium oxysporum f. sp. cepae. Sci. Rep. 2018;8:13530. doi: 10.1038/s41598-018-30335-7. - DOI - PMC - PubMed
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

