Whole Genome Sequencing in the Evaluation of Fetal Structural Anomalies: A Parallel Test with Chromosomal Microarray Plus Whole Exome Sequencing
- PMID: 33800913
- PMCID: PMC7999180
- DOI: 10.3390/genes12030376
Whole Genome Sequencing in the Evaluation of Fetal Structural Anomalies: A Parallel Test with Chromosomal Microarray Plus Whole Exome Sequencing
Abstract
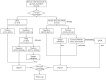
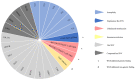
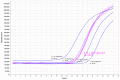
Whole genome sequencing (WGS) is a powerful tool for postnatal genetic diagnosis, but relevant clinical studies in the field of prenatal diagnosis are limited. The present study aimed to prospectively evaluate the utility of WGS compared with chromosomal microarray (CMA) and whole exome sequencing (WES) in the prenatal diagnosis of fetal structural anomalies. We performed trio WGS (≈40-fold) in parallel with CMA in 111 fetuses with structural or growth anomalies, and sequentially performed WES when CMA was negative (CMA plus WES). In comparison, WGS not only detected all pathogenic genetic variants in 22 diagnosed cases identified by CMA plus WES, yielding a diagnostic rate of 19.8% (22/110), but also provided additional and clinically significant information, including a case of balanced translocations and a case of intrauterine infection, which might not be detectable by CMA or WES. WGS also required less DNA (100 ng) as input and could provide a rapid turnaround time (TAT, 18 ± 6 days) compared with that (31 ± 8 days) of the CMA plus WES. Our results showed that WGS provided more comprehensive and precise genetic information with a rapid TAT and less DNA required than CMA plus WES, which enables it as an alternative prenatal diagnosis test for fetal structural anomalies.
Keywords: chromosomal microarray; fetal structural anomalies; prenatal diagnosis; whole exome sequencing; whole genome sequencing.
Conflict of interest statement
Z.Y., J.S., L.L., F.L., S.C., X.L., X.X.W., Z.W., Y.W., S.Y., F.Z., and Z.P. are current employees of BGI Genomics. All other authors declare that they have no financial interests.
Figures

Similar articles
-
Comprehensive prenatal diagnostics: Exome versus genome sequencing.Prenat Diagn. 2023 Aug;43(9):1132-1141. doi: 10.1002/pd.6402. Epub 2023 Jul 3. Prenat Diagn. 2023. PMID: 37355983
-
Whole-exome sequencing in the evaluation of fetal structural anomalies: a prospective cohort study.Lancet. 2019 Feb 23;393(10173):758-767. doi: 10.1016/S0140-6736(18)32042-7. Epub 2019 Jan 31. Lancet. 2019. PMID: 30712878
-
Prenatal genetic diagnosis of omphalocele by karyotyping, chromosomal microarray analysis and exome sequencing.Ann Med. 2021 Dec;53(1):1285-1291. doi: 10.1080/07853890.2021.1962966. Ann Med. 2021. PMID: 34374610 Free PMC article.
-
Whole exome sequencing in fetuses with isolated increased nuchal translucency: a systematic review and meta-analysis.J Matern Fetal Neonatal Med. 2023 Dec;36(1):2193285. doi: 10.1080/14767058.2023.2193285. J Matern Fetal Neonatal Med. 2023. PMID: 37019452
-
Prenatal Genetic Diagnosis of Fetal Cystic Hygroma: A Retrospective Single-Center Study from China.Cytogenet Genome Res. 2022;162(7):354-364. doi: 10.1159/000528600. Epub 2023 Mar 10. Cytogenet Genome Res. 2022. PMID: 36907182 Review.
Cited by
-
Molecular Approaches in Fetal Malformations, Dynamic Anomalies and Soft Markers: Diagnostic Rates and Challenges-Systematic Review of the Literature and Meta-Analysis.Diagnostics (Basel). 2022 Feb 23;12(3):575. doi: 10.3390/diagnostics12030575. Diagnostics (Basel). 2022. PMID: 35328129 Free PMC article. Review.
-
Prenatal diagnosis of mucopolysaccharidosis type I on hepatosplenomegaly and coarse features: a case-report.BMC Pregnancy Childbirth. 2025 Jan 3;25(1):3. doi: 10.1186/s12884-024-07115-5. BMC Pregnancy Childbirth. 2025. PMID: 39754079 Free PMC article.
-
A case of Aicardi-Goutières syndrome caused by TREX1 gene mutation.BMC Pregnancy Childbirth. 2023 Feb 22;23(1):124. doi: 10.1186/s12884-023-05436-5. BMC Pregnancy Childbirth. 2023. PMID: 36814213 Free PMC article.
-
Hydrocephalus and Growth Retardation: A Fetal RNU4ATAC-opathy Missed by Whole-Exome Sequencing.Mol Syndromol. 2023 Jan;13(6):522-526. doi: 10.1159/000524501. Epub 2022 May 9. Mol Syndromol. 2023. PMID: 36660028 Free PMC article.
-
The Value of a Comprehensive Genomic Evaluation in Prenatal Diagnosis of Genetic Diseases: A Retrospective Study.Genes (Basel). 2022 Dec 14;13(12):2365. doi: 10.3390/genes13122365. Genes (Basel). 2022. PMID: 36553632 Free PMC article.
References
-
- Callaway J.L., Shaffer L.G., Chitty L.S., Rosenfeld J.A., Crolla J.A. The clinical utility of microarray technologies applied to prenatal cytogenetics in the presence of a normal conventional karyotype: A review of the literature. Prenat. Diagn. 2013;33:1119–1123. doi: 10.1002/pd.4209. - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

