Genome Sequencing of five Lacticaseibacillus Strains and Analysis of Type I and II Toxin-Antitoxin System Distribution
- PMID: 33800997
- PMCID: PMC8003834
- DOI: 10.3390/microorganisms9030648
Genome Sequencing of five Lacticaseibacillus Strains and Analysis of Type I and II Toxin-Antitoxin System Distribution
Abstract
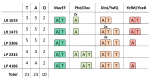
The analysis of bacterial genomes is a potent tool to investigate the distribution of specific traits related to the ability of surviving in particular environments. Among the traits associated with the adaptation to hostile conditions, toxin-antitoxin (TA) systems have recently gained attention in lactic acid bacteria. In this work, genome sequences of Lacticaseibacillus strains of dairy origin were compared, focusing on the distribution of type I TA systems homologous to Lpt/RNAII and of the most common type II TA systems. A high number of TA systems have been identified spread in all the analyzed strains, with type I TA systems mainly located on plasmid DNA. The type II TA systems identified in these strains highlight the diversity of encoded toxins and antitoxins and their organization. This study opens future perspectives on the use of genomic data as a resource for the study of TA systems distribution and prevalence in microorganisms of industrial relevance.
Keywords: DinJ/YafQ; Lacticaseibacillus; Lpt toxin; MazEF; Phd/Doc; YefM/YoeB; genome sequencing; toxin-antitoxin systems.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures



Similar articles
-
Functional characterization and transcriptional repression by Lacticaseibacillus paracasei DinJ-YafQ.Appl Microbiol Biotechnol. 2022 Nov;106(21):7113-7128. doi: 10.1007/s00253-022-12195-4. Epub 2022 Oct 4. Appl Microbiol Biotechnol. 2022. PMID: 36194262 Free PMC article.
-
Edwardsiella piscicida YefM-YoeB: A Type II Toxin-Antitoxin System That Is Related to Antibiotic Resistance, Biofilm Formation, Serum Survival, and Host Infection.Front Microbiol. 2021 Mar 1;12:646299. doi: 10.3389/fmicb.2021.646299. eCollection 2021. Front Microbiol. 2021. PMID: 33732226 Free PMC article.
-
Expression of DinJ-YafQ System of Lactobacillus casei Group Strains in Response to Food Processing Stresses.Microorganisms. 2019 Oct 11;7(10):438. doi: 10.3390/microorganisms7100438. Microorganisms. 2019. PMID: 31614503 Free PMC article.
-
Keeping the Wolves at Bay: Antitoxins of Prokaryotic Type II Toxin-Antitoxin Systems.Front Mol Biosci. 2016 Mar 22;3:9. doi: 10.3389/fmolb.2016.00009. eCollection 2016. Front Mol Biosci. 2016. PMID: 27047942 Free PMC article. Review.
-
Evaluating the Potential for Cross-Interactions of Antitoxins in Type II TA Systems.Toxins (Basel). 2020 Jun 26;12(6):422. doi: 10.3390/toxins12060422. Toxins (Basel). 2020. PMID: 32604745 Free PMC article. Review.
Cited by
-
Functional characterization and transcriptional repression by Lacticaseibacillus paracasei DinJ-YafQ.Appl Microbiol Biotechnol. 2022 Nov;106(21):7113-7128. doi: 10.1007/s00253-022-12195-4. Epub 2022 Oct 4. Appl Microbiol Biotechnol. 2022. PMID: 36194262 Free PMC article.
-
Lactic acid bacteria in cow raw milk for cheese production: Which and how many?Front Microbiol. 2023 Jan 12;13:1092224. doi: 10.3389/fmicb.2022.1092224. eCollection 2022. Front Microbiol. 2023. PMID: 36713157 Free PMC article. Review.
-
Strategies to Investigate Membrane Damage, Nucleoid Condensation, and RNase Activity of Bacterial Toxin-Antitoxin Systems.Methods Protoc. 2021 Oct 8;4(4):71. doi: 10.3390/mps4040071. Methods Protoc. 2021. PMID: 34698227 Free PMC article.
-
Lacticaseibacillus Strains Isolated from Raw Milk: Screening Strategy for Their Qualification as Adjunct Culture in Cheesemaking.Foods. 2023 Oct 29;12(21):3949. doi: 10.3390/foods12213949. Foods. 2023. PMID: 37959068 Free PMC article.
-
Activity of Membrane-Permeabilizing Lpt Peptides.Biomolecules. 2024 Aug 13;14(8):994. doi: 10.3390/biom14080994. Biomolecules. 2024. PMID: 39199383 Free PMC article.
References
-
- Mohammadzadeh R., Shivaee A., Ohadi E., Kalani B.S. In Silico Insight into the Dominant Type II Toxin–Antitoxin Systems and Clp Proteases in Listeria monocytogenes and Designation of Derived Peptides as a Novel Approach to Interfere with this System. Int. J. Pept. Res. Ther. 2020;26:613–623. doi: 10.1007/s10989-019-09868-6. - DOI
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

