Insulin-Like Growth Factor Binding Protein-3 Exerts Its Anti-Metastatic Effect in Aerodigestive Tract Cancers by Disrupting the Protein Stability of Vimentin
- PMID: 33801272
- PMCID: PMC7958122
- DOI: 10.3390/cancers13051041
Insulin-Like Growth Factor Binding Protein-3 Exerts Its Anti-Metastatic Effect in Aerodigestive Tract Cancers by Disrupting the Protein Stability of Vimentin
Abstract
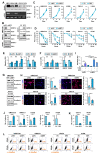
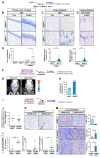
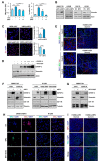
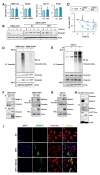
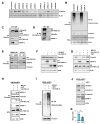
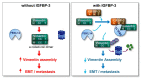
The proapoptotic, antiangiogenic, and antimetastatic activities of insulin-like growth factor binding protein-3 (IGFBP-3) through IGF-dependent or -independent mechanisms have been suggested in various types of human cancers. However, a mechanistic explanation of and downstream targets involved in the antimetastatic effect of IGFBP-3 is still lacking. In this study, by applying various in vitro and in vivo models, we show that IGFBP-3 suppresses migration and invasion of human head and neck squamous carcinoma (HNSCC) and non-small cell lung cancer (NSCLC) cells. Silencing IGFBP-3 expression elevated the migration and invasion of NSCLC and HNSCC cells in vitro and their local invasion and metastasis in vivo, whereas overexpression of IGFBP-3 decreased such prometastatic changes. Local invasion of 4-nitroquinoline-1-oxide (4-NQO)-induced HNSCC tumors was consistently significantly potentiated in Igfbp3 knockout mice compared with that in wild-type mice. Mechanistically, IGFBP-3 disrupted the protein stability of vimentin via direct binding and promoting its association with the E3 ligase FBXL14, causing proteasomal degradation. The C-terminal domain of IGFBP-3 and the head domain of vimentin are essential for their interaction. These results provide a molecular framework for IGFBP-3's IGF-independent antimetastatic and antitumor activities.
Keywords: insulin-like growth factor binding protein-3; metastasis; vimentin.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

