Distribution of Extended-Spectrum β-Lactamase (ESBL)-Encoding Genes among Multidrug-Resistant Gram-Negative Pathogens Collected from Three Different Countries
- PMID: 33801418
- PMCID: PMC7998439
- DOI: 10.3390/antibiotics10030247
Distribution of Extended-Spectrum β-Lactamase (ESBL)-Encoding Genes among Multidrug-Resistant Gram-Negative Pathogens Collected from Three Different Countries
Abstract
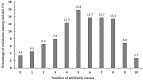
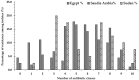
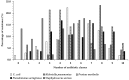
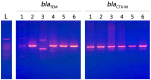
The incidence of Extended-spectrum β-lactamase (ESBL)-encoding genes (blaCTX-M and blaTEM) among Gram-negative multidrug-resistant pathogens collected from three different countries was investigated. Two hundred and ninety-two clinical isolates were collected from Egypt (n = 90), Saudi Arabia (n = 162), and Sudan (n = 40). Based on the antimicrobial sensitivity against 20 antimicrobial agents from 11 antibiotic classes, the most resistant strains were selected and identified using the Vitek2 system and 16S rRNA gene sequence analysis. A total of 85.6% of the isolates were found to be resistant to more than three antibiotic classes. The ratios of the multidrug-resistant strains for Egypt, Saudi Arabia, and Sudan were 74.4%, 90.1%, and 97.5%, respectively. Escherichia coli, Klebsiella pneumoniae, and Pseudomonas aeruginosa showed inconstant resistance levels to the different classes of antibiotics. Escherichia coli and Klebsiella pneumoniae had the highest levels of resistance against macrolides followed by penicillins and cephalosporin, while Pseudomonas aeruginosa was most resistant to penicillins followed by classes that varied among different countries. The isolates were positive for the presence of the blaCTX-M and blaTEM genes. The blaCTX-M gene was the predominant gene in all isolates (100%), while blaTEM was detected in 66.7% of the selected isolates. This work highlights the detection of multidrug-resistant bacteria and resistant genes among different countries. We suggest that the medical authorities urgently implement antimicrobial surveillance plans and infection control policies for early detection and effective prevention of the rapid spread of these pathogens.
Keywords: antimicrobial resistance; clinical samples; different countries; multidrug-resistant pathogens; resistant-genes.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Sahm D.F., Thornsberry C., Mayfield D.C., Jones M.E., Karlowsky J.A. Multidrug-resistant urinary tract isolates of Escherichia coli: Prevalence and patient demographics in the United States in 2000. Antimicrob. Agents Chemother. 2001;45:1402–1406. doi: 10.1128/AAC.45.5.1402-1406.2001. - DOI - PMC - PubMed
-
- Karlowsky J.A., Jones M.E., Draghi D.C., Thornsberry C., Sahm D.F., Volturo G.A. Prevalence and antimicrobial susceptibilities of bacteria isolated from blood cultures of hospitalized patients in the United States in 2002. Ann. Clin. Microbiol. Antimicrob. 2004;3:7. doi: 10.1186/1476-0711-3-7. - DOI - PMC - PubMed
-
- Van De Sande-Bruinsma N., Grundmann H., Verloo D., Tiemersma E., Monen J., Goossens H., Ferech M., European Antimicrobial Resistance Surveillance System Group. European Surveillance of Antimicrobial Consumption Project Group Antimicrobial drug use and resistance in Europe. Emerg. Infect. Dis. 2008;14:1722–1730. doi: 10.3201/eid1411.070467. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

