Infection of Chinese Rhesus Monkeys with a Subtype C SHIV Resulted in Attenuated In Vivo Viral Replication Despite Successful Animal-to-Animal Serial Passages
- PMID: 33801437
- PMCID: PMC7998229
- DOI: 10.3390/v13030397
Infection of Chinese Rhesus Monkeys with a Subtype C SHIV Resulted in Attenuated In Vivo Viral Replication Despite Successful Animal-to-Animal Serial Passages
Abstract
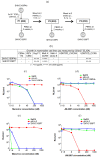
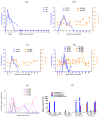
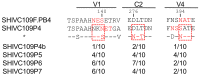
Rhesus macaques can be readily infected with chimeric simian-human immunodeficiency viruses (SHIV) as a suitable virus challenge system for testing the efficacy of HIV vaccines. Three Chinese-origin rhesus macaques (ChRM) were inoculated intravenously (IV) with SHIVC109P4 in a rapid serial in vivo passage. SHIV recovered from the peripheral blood of the final ChRM was used to generate a ChRM-adapted virus challenge stock. This stock was titrated for the intrarectal route (IR) in 8 ChRMs using undiluted, 1:10 or 1:100 dilutions, to determine a suitable dose for use in future vaccine efficacy testing via repeated low-dose IR challenges. All 11 ChRMs were successfully infected, reaching similar median peak viraemias at 1-2 weeks post inoculation but undetectable levels by 8 weeks post inoculation. T-cell responses were detected in all animals and Tier 1 neutralizing antibodies (Nab) developed in 10 of 11 infected ChRMs. All ChRMs remained healthy and maintained normal CD4+ T cell counts. Sequence analyses showed >98% amino acid identity between the original inoculum and virus recovered at peak viraemia indicating only minimal changes in the env gene. Thus, while replication is limited over time, our adapted SHIV can be used to test for protection of virus acquisition in ChRMs.
Keywords: Chinese rhesus macaques; SHIV; subtype C.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures


References
-
- Nishimura Y., Igarashi T., Donau O.K., Buckler-White A., Buckler C., Lafont B.A., Goeken R.M., Goldstein S., Hirsch V.M., Martin M.A. Highly pathogenic SHIVs and SIVs target different CD4+ T cell subsets in rhesus monkeys, explaining their divergent clinical courses. Proc. Natl. Acad. Sci. USA. 2004;101:12324–12329. doi: 10.1073/pnas.0404620101. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

