Gene Transactivation and Transrepression in MYC-Driven Cancers
- PMID: 33801599
- PMCID: PMC8037706
- DOI: 10.3390/ijms22073458
Gene Transactivation and Transrepression in MYC-Driven Cancers
Abstract
MYC is a proto-oncogene regulating a large number of genes involved in a plethora of cellular functions. Its deregulation results in activation of MYC gene expression and/or an increase in MYC protein stability. MYC overexpression is a hallmark of malignant growth, inducing self-renewal of stem cells and blocking senescence and cell differentiation. This review summarizes the latest advances in our understanding of MYC-mediated molecular mechanisms responsible for its oncogenic activity. Several recent findings indicate that MYC is a regulator of cancer genome and epigenome: MYC modulates expression of target genes in a site-specific manner, by recruiting chromatin remodeling co-factors at promoter regions, and at genome-wide level, by regulating the expression of several epigenetic modifiers that alter the entire chromatin structure. We also discuss novel emerging therapeutic strategies based on both direct modulation of MYC and its epigenetic cofactors.
Keywords: MYC; MYC deregulation; MYC-driven cancers; epigenetic modulation; therapeutic target; therapy resistance.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
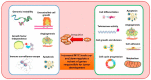
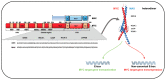
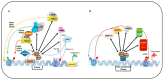
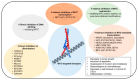
References
-
- Finver S.N., Nishikura K., Finger L.R., Haluska F.G., Finan J., Nowell P.C., Croce C.M. Sequence analysis of the MYC oncogene involved in the t(8;14)(q24;q11) chromosome translocation in a human leukemia T-cell line indicates that putative regulatory regions are not altered. Proc. Natl. Acad. Sci. USA. 1988;85:3052–3056. doi: 10.1073/pnas.85.9.3052. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

