Laser Microdissection of Specific Stem-Base Tissue Types from Olive Microcuttings for Isolation of High-Quality RNA
- PMID: 33801829
- PMCID: PMC7999021
- DOI: 10.3390/biology10030209
Laser Microdissection of Specific Stem-Base Tissue Types from Olive Microcuttings for Isolation of High-Quality RNA
Erratum in
-
Erratum: Velada et al. Laser Microdissection of Specific Stem-Base Tissue Types from Olive Microcuttings for Isolation of High-Quality RNA. Biology 2021, 10, 209.Biology (Basel). 2021 Jul 28;10(8):718. doi: 10.3390/biology10080718. Biology (Basel). 2021. PMID: 34440052 Free PMC article.
Abstract
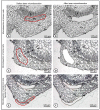
Higher plants are composed of different tissue and cell types. Distinct cells host different biochemical and physiological processes which is reflected in differences in gene expression profiles, protein and metabolite levels. When omics are to be carried out, the information provided by a specific cell type can be diluted and/or masked when using a mixture of distinct cells. Thus, studies performed at the cell- and tissue-type level are gaining increasing interest. Laser microdissection (LM) technology has been used to isolate specific tissue and cell types. However, this technology faces some challenges depending on the plant species and tissue type under analysis. Here, we show for the first time a LM protocol that proved to be efficient for harvesting specific tissue types (phloem, cortex and epidermis) from olive stem nodal segments and obtaining RNA of high quality. This is important for future transcriptomic studies to identify rooting-competent cells. Here, nodal segments were flash-frozen in liquid nitrogen-cooled isopentane and cryosectioned. Albeit the lack of any fixatives used to preserve samples' anatomy, cryosectioned sections showed tissues with high morphological integrity which was comparable with that obtained with the paraffin-embedding method. Cells from the phloem, cortex and epidermis could be easily distinguished and efficiently harvested by LM. Total RNA isolated from these tissues exhibited high quality with RNA Quality Numbers (determined by a Fragment Analyzer System) ranging between 8.1 and 9.9. This work presents a simple, rapid and efficient LM procedure for harvesting specific tissue types of olive stems and obtaining high-quality RNA.
Keywords: OCT medium; PEN-membrane glass slide; RNA quality number; adventitious roots; cortex; cryosectioning; epidermis; phloem; single cell.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
A method for obtaining high quality RNA from paraffin sections of plant tissues by laser microdissection.J Plant Res. 2010 Nov;123(6):807-13. doi: 10.1007/s10265-010-0319-4. Epub 2010 Mar 11. J Plant Res. 2010. PMID: 20221666
-
Developing a simple and rapid method for cell-specific transcriptome analysis through laser microdissection: insights from citrus rind with broader implications.Plant Methods. 2024 Jul 27;20(1):113. doi: 10.1186/s13007-024-01242-y. Plant Methods. 2024. PMID: 39068421 Free PMC article.
-
Application of laser microdissection to study plant-fungal pathogen interactions.Methods Mol Biol. 2010;638:153-63. doi: 10.1007/978-1-60761-611-5_11. Methods Mol Biol. 2010. PMID: 20238267
-
Laser microdissection of plant tissue: what you see is what you get.Annu Rev Plant Biol. 2006;57:181-201. doi: 10.1146/annurev.arplant.56.032604.144138. Annu Rev Plant Biol. 2006. PMID: 16669760 Review.
-
Complementary techniques: laser capture microdissection--increasing specificity of gene expression profiling of cancer specimens.Adv Exp Med Biol. 2007;593:54-65. doi: 10.1007/978-0-387-39978-2_6. Adv Exp Med Biol. 2007. PMID: 17265716 Review.
Cited by
-
Erratum: Velada et al. Laser Microdissection of Specific Stem-Base Tissue Types from Olive Microcuttings for Isolation of High-Quality RNA. Biology 2021, 10, 209.Biology (Basel). 2021 Jul 28;10(8):718. doi: 10.3390/biology10080718. Biology (Basel). 2021. PMID: 34440052 Free PMC article.
-
Stimulation of adventitious root formation by laser wounding in rose cuttings: A matter of energy and pattern.Front Plant Sci. 2022 Sep 29;13:1009085. doi: 10.3389/fpls.2022.1009085. eCollection 2022. Front Plant Sci. 2022. PMID: 36247617 Free PMC article.
References
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

