Direct Flavonoid-Focused Chemical Comparison among Three Epimedium Plants by Online Liquid Extraction-High Performance Liquid Chromatography-Tandem Mass Spectrometry
- PMID: 33802139
- PMCID: PMC7998785
- DOI: 10.3390/molecules26061520
Direct Flavonoid-Focused Chemical Comparison among Three Epimedium Plants by Online Liquid Extraction-High Performance Liquid Chromatography-Tandem Mass Spectrometry
Abstract
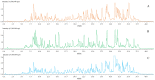
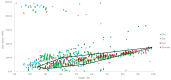
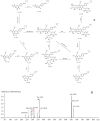
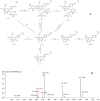
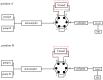
It is usually a tedious task to profile the chemical composition of a given herbal medicine (HM) using high performance liquid chromatography-tandem mass spectrometry (LC-MS/MS) due to the time-consuming sample preparation and laborious post-acquisition data processing procedures. Even worse, some labile compounds may face degradation risks when exposed to organic solvents for a relatively long period. As one of the most popular HMs, the promising therapeutic benefits of Epimedii Herba (Chinese name: Yinyanghuo) are well defined; however, the chemical profile, and in particular those flavonoids that have been claimed to be responsible for the efficacy, remains largely unknown. Attempts are devoted here to achieve direct LC-MS measurement and efficient post-acquisition data processing, and chemome comparison among three original sources of Epimedii Herba, such as Epimedium sagittatum (Esa), E. pubescens (Epu), and E. koreanum (Eko) was employed to illustrate the strategy utility. A home-made online liquid extraction (OLE) module was introduced at the front of the analytical column to comprehensively transfer the compounds from raw materials onto the LC-MS instrument. A mass defect filtering approach was programmed to efficiently mine the massive LC-MS dataset after which a miniature database was built involving all chemical information of flavonoids from the genus Epimedium to draw a pentagonal frame to rapidly capture potential quasi-molecular ions (mainly [M-H]-). A total of 99 flavonoids (66 in Esa, 84 in Eko, and 66 in Epu) were captured, and structurally annotated by summarizing the mass fragmentation pathways from the mass spectrometric data of authentic compounds and an in-house data library as well. Noteworthily, neutral loss of 144 Da was firstly assigned to the neutral cleavage of rhamnosyl residues. Significant species-differences didn't occur among their chemical patterns. The current study proposed a robust strategy enabling rapid chemical profiling of, but not limited to, HMs.
Keywords: Epimedii Herba; flavonoids; mass defect filtering; neutral loss of rhamnosyl residue; online liquid extraction.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Direct infusion-tandem mass spectrometry combining with data mining strategies enables rapid chemome characterization of medicinal plants: A case study of Polygala tenuifolia.J Pharm Biomed Anal. 2021 Sep 10;204:114281. doi: 10.1016/j.jpba.2021.114281. Epub 2021 Jul 24. J Pharm Biomed Anal. 2021. PMID: 34333452
-
[Chemome profiling of Pien-Tze-Huang by online pressurized liquid extraction-ultra-high performance liquid chromatography-ion trap-time-of-flight mass spectrometry].Se Pu. 2021 May;39(5):478-487. doi: 10.3724/SP.J.1123.2020.10011. Se Pu. 2021. PMID: 34227332 Free PMC article. Chinese.
-
A novel online preparative high-performance liquid chromatography system with the multiple trap columns-valve switch technique for the rapid and efficient isolation of main flavonoids from Epimedium koreanum Nakai.J Sep Sci. 2021 Jan;44(2):656-665. doi: 10.1002/jssc.202000783. Epub 2020 Nov 27. J Sep Sci. 2021. PMID: 33151025
-
Online pressurized liquid extraction enables directly chemical analysis of herbal medicines: A mini review.J Pharm Biomed Anal. 2021 Oct 25;205:114332. doi: 10.1016/j.jpba.2021.114332. Epub 2021 Aug 19. J Pharm Biomed Anal. 2021. PMID: 34455204 Review.
-
[A review on research of raw material and cut crude drug of Herba epimedii in last ten years].Zhongguo Zhong Yao Za Zhi. 2007 Mar;32(6):466-71. Zhongguo Zhong Yao Za Zhi. 2007. PMID: 17552145 Review. Chinese.
Cited by
-
Online Extraction Followed by LC-MS/MS Analysis of Lipids in Natural Samples: A Proof-of-Concept Profiling Lecithin in Seeds.Foods. 2023 Jan 7;12(2):281. doi: 10.3390/foods12020281. Foods. 2023. PMID: 36673373 Free PMC article.
-
Phytochemical Profile and Antioxidant Activities of Coleus amboinicus Lour. Cultivated in Indonesia and Poland.Molecules. 2021 May 14;26(10):2915. doi: 10.3390/molecules26102915. Molecules. 2021. PMID: 34068950 Free PMC article.
-
Aqueous Extracts of Ocimum gratissimum Sensitize Hepatocellular Carcinoma Cells to Cisplatin through BRCA1 Inhibition.Int J Mol Sci. 2024 Aug 1;25(15):8424. doi: 10.3390/ijms25158424. Int J Mol Sci. 2024. PMID: 39125994 Free PMC article.
References
-
- Wang H.D., Zhang C.X., Zuo T.T., Li W.W., Li J., Wang X.Y., Qian Y.X., Guo D.A., Yang W.Z. In–depth profiling, characterization, and comparison of the ginsenosides among three different parts (the root, stem leaf, and flower bud) of Panax quinquefolius L. by ultra–high performance liquid chromatography/quadrupole–Orbitrap mass spectrometry. Anal. Bioanal. Chem. 2019;411:7817–7829. doi: 10.1007/s00216-019-02180-8. - DOI - PubMed
-
- Yang W.Z., Shi X.J., Yao C.L., Huang Y., Hou J.J., Han S.M., Feng Z.J., Wei W.L., Wu W.Y., Guo D.A. A novel neutral loss/product ion scan–incorporated integral approach for the untargeted characterization and comparison of the carboxyl–free ginsenosides from Panax ginseng, Panax quinquefolius, and Panax notoginseng. J. Pharm. Biomed. Anal. 2020;177:112813. doi: 10.1016/j.jpba.2019.112813. - DOI - PubMed
-
- Shi X.J., Yang W.Z., Huang Y., Hou J.J., Qiu S., Yao C.L., Feng Z.J., Wei W.L., Wu W.Y., Guo D.A. Direct screening of malonylginsenosides from nine Ginseng extracts by an untargeted profiling strategy incorporating in–source collision–induced dissociation, mass tag, and neutral loss scan on a hybrid linear ion–trap/Orbitrap mass spectrometer coupled to ultra–high performance liquid chromatography. J. Chromatogr. A. 2018;1571:213–222. - PubMed
-
- Song Y.L., Zhang N., Shi S.P., Li J., Zhao Y.F., Zhang Q., Jiang Y., Tu P.F. Homolog–focused profiling of ginsenosides based on the integration of step–wise formate anion–to–deprotonated ion transition screening and scheduled multiple reaction monitoring. J. Chromatogr. A. 2015;1406:136–144. doi: 10.1016/j.chroma.2015.06.007. - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

