The Tumor Proteolytic Landscape: A Challenging Frontier in Cancer Diagnosis and Therapy
- PMID: 33802262
- PMCID: PMC7958950
- DOI: 10.3390/ijms22052514
The Tumor Proteolytic Landscape: A Challenging Frontier in Cancer Diagnosis and Therapy
Abstract
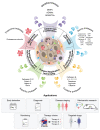
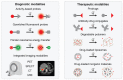
In recent decades, dysregulation of proteases and atypical proteolysis have become increasingly recognized as important hallmarks of cancer, driving community-wide efforts to explore the proteolytic landscape of oncologic disease. With more than 100 proteases currently associated with different aspects of cancer development and progression, there is a clear impetus to harness their potential in the context of oncology. Advances in the protease field have yielded technologies enabling sensitive protease detection in various settings, paving the way towards diagnostic profiling of disease-related protease activity patterns. Methods including activity-based probes and substrates, antibodies, and various nanosystems that generate reporter signals, i.e., for PET or MRI, after interaction with the target protease have shown potential for clinical translation. Nevertheless, these technologies are costly, not easily multiplexed, and require advanced imaging technologies. While the current clinical applications of protease-responsive technologies in oncologic settings are still limited, emerging technologies and protease sensors are poised to enable comprehensive exploration of the tumor proteolytic landscape as a diagnostic and therapeutic frontier. This review aims to give an overview of the most relevant classes of proteases as indicators for tumor diagnosis, current approaches to detect and monitor their activity in vivo, and associated therapeutic applications.
Keywords: protease activity; protease diagnostic and therapeutic modalities; tumor microenvironment.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
-
- Rawlings N.D., Barrett A.J., Thomas P.D., Huang X., Bateman A., Finn R.D. The MEROPS database of proteolytic enzymes, their substrates and inhibitors in 2017 and a comparison with peptidases in the PANTHER database. Nucleic Acids Res. 2018;46:D624–D632. doi: 10.1093/nar/gkx1134. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

