Site-Specific Fluorescent Labeling of RNA Interior Positions
- PMID: 33802273
- PMCID: PMC7959133
- DOI: 10.3390/molecules26051341
Site-Specific Fluorescent Labeling of RNA Interior Positions
Abstract
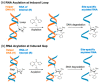
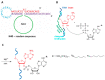
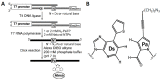
The introduction of fluorophores into RNA for both in vitro and in cellulo studies of RNA function and cellular distribution is a subject of great current interest. Here I briefly review methods, some well-established and others newly developed, which have been successfully exploited to site-specifically fluorescently label interior positions of RNAs, as a guide to investigators seeking to apply this approach to their studies. Most of these methods can be applied directly to intact RNAs, including (1) the exploitation of natural posttranslational modifications, (2) the repurposing of enzymatic transferase reactions, and (3) the nucleic acid-assisted labeling of intact RNAs. In addition, several methods are described in which specifically labeled RNAs are prepared de novo.
Keywords: RNA interior positions; site-specific fluorescent labeling.
Conflict of interest statement
The author declares no conflict of interest.
Figures



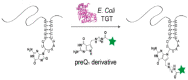


References
-
- Saito Y., Hudson R.H. Base-modified fluorescent purine nucleosides and nucleotides for use in oligonucleotide probes. J. Photochem. Photobiol. C. Photochem. Rev. 2018;36:48–73. doi: 10.1016/j.jphotochemrev.2018.07.001. - DOI
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

