EphB6 Regulates TFEB-Lysosomal Pathway and Survival of Disseminated Indolent Breast Cancer Cells
- PMID: 33802447
- PMCID: PMC7959459
- DOI: 10.3390/cancers13051079
EphB6 Regulates TFEB-Lysosomal Pathway and Survival of Disseminated Indolent Breast Cancer Cells
Abstract
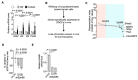
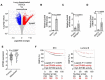
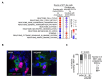
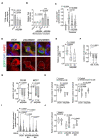
Late relapse of disseminated cancer cells is a common feature of breast and prostate tumors. Several intrinsic and extrinsic factors have been shown to affect quiescence and reawakening of disseminated dormant cancer cells (DDCCs); however, the signals and processes sustaining the survival of DDCCs in a foreign environment are still poorly understood. We have recently shown that crosstalk with lung epithelial cells promotes survival of DDCCs of estrogen receptor-positive (ER+) breast tumors. By using a lung organotypic system and in vivo dissemination assays, here we show that the TFEB-lysosomal axis is activated in DDCCs and that it is modulated by the pro-survival ephrin receptor EphB6. TFEB lysosomal direct targets are enriched in DDCCs in vivo and correlate with relapse in ER+ breast cancer patients. Direct coculture of DDCCs with alveolar type I-like lung epithelial cells and dissemination in the lung drive lysosomal accumulation and EphB6 induction. EphB6 contributes to survival, TFEB transcriptional activity, and lysosome formation in DDCCs in vitro and in vivo. Furthermore, signaling from EphB6 promotes the proliferation of surrounding lung parenchymal cells in vivo. Our data provide evidence that EphB6 is a key factor in the crosstalk between disseminated dormant cancer cells and the lung parenchyma and that the TFEB-lysosomal pathway plays an important role in the persistence of DDCCs.
Keywords: EphB6; Ephrin receptors; breast cancer; dormancy; lysosomes; metastasis; tumor microenvironment.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Gawrzak S., Rinaldi L., Gregorio S., Arenas E.J., Salvador F., Urosevic J., Figueras-Puig C., Rojo F., Del Barco Barrantes I., Cejalvo J.M., et al. MSK1 Regulates Luminal Cell Differentiation and Metastatic Dormancy in ER + Breast Cancer. Nat. Cell Biol. 2018;20:211–221. doi: 10.1038/s41556-017-0021-z. - DOI - PubMed
-
- Aguirre-Ghiso J.A., Sosa M.S. Emerging Topics on Disseminated Cancer Cell Dormancy and the Paradigm of Metastasis. Annual Rev. Cancer Biol. 2018;2:377–393. doi: 10.1146/annurev-cancerbio-030617-050446. - DOI
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

