A Streamlined Approach to Pathway Analysis from RNA-Sequencing Data
- PMID: 33802642
- PMCID: PMC8006023
- DOI: 10.3390/mps4010021
A Streamlined Approach to Pathway Analysis from RNA-Sequencing Data
Abstract
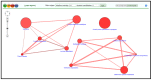
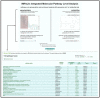
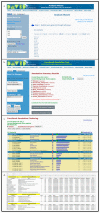
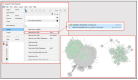
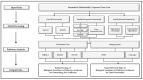
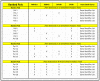
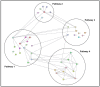
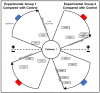
The reduction in costs associated with performing RNA-sequencing has driven an increase in the application of this analytical technique; however, restrictive factors associated with this tool have now shifted from budgetary constraints to time required for data processing. The sheer scale of the raw data produced can present a formidable challenge for researchers aiming to glean vital information about samples. Though many of the companies that perform RNA-sequencing provide a basic report for the submitted samples, this may not adequately capture particular pathways of interest for sample comparisons. To further assess these data, it can therefore be necessary to utilize various enrichment and mapping software platforms to highlight specific relations. With the wide array of these software platforms available, this can also present a daunting task. The methodology described herein aims to enable researchers new to handling RNA-sequencing data with a streamlined approach to pathway analysis. Additionally, the implemented software platforms are readily available and free to utilize, making this approach viable, even for restrictive budgets. The resulting tables and nodal networks will provide valuable insight into samples and can be used to generate high-quality graphics for publications and presentations.
Keywords: RNA-sequencing; cytoscape; data processing; database; enrichment analysis; mapping; network; protocol; transcriptomics.
Conflict of interest statement
The author declares no conflict of interest.
Figures


References
-
- Afgan E., Baker D., Batut B., van den Beek M., Bouvier D., Čech M., Chilton J., Clements D., Coraor N., Gruning B.A., et al. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2018 update. Nucleic Acids Res. 2018;46:W537–W544. doi: 10.1093/nar/gky379. - DOI - PMC - PubMed
-
- Torson A.S., Dong Y.-W., Sinclair B.J. Help, there are ‘omics’ in my comparative physiology! J. Exp. Biol. 2020;223:191262. - PubMed
-
- Chen L., Fei C., Zhu L., Xu Z., Zou W., Yang T., Lin H., Xi D. RNA-seq approach to analysis of gene expression profiles in dark green islands and light green tissues of Cucumber mosaic virus-infected Nicotiana tabacum. PLoS ONE. 2017;12:e0175391. doi: 10.1371/journal.pone.0175391. - DOI - PMC - PubMed
-
- Warden C.D., Yuan Y.-C., Wu X. Optimal calculation of RNA-Seq fold-change values. Int. J. Comput. Bioinform. Silico Model. 2013;2:285–292.
LinkOut - more resources
Full Text Sources
Other Literature Sources

