Recent Updates on the Significance of KRAS Mutations in Colorectal Cancer Biology
- PMID: 33802849
- PMCID: PMC8002639
- DOI: 10.3390/cells10030667
Recent Updates on the Significance of KRAS Mutations in Colorectal Cancer Biology
Abstract
The most commonly mutated isoform of RAS among all cancer subtypes is KRAS. In this review, we focus on the special role of KRAS mutations in colorectal cancer (CRC), aiming to collect recent data on KRAS-driven enhanced cell signalling, in vitro and in vivo research models, and CRC development-related processes such as metastasis and cancer stem cell formation. We attempt to cover the diverse nature of the effects of KRAS mutations on age-related CRC development. As the incidence of CRC is rising in young adults, we have reviewed the driving forces of ageing-dependent CRC.
Keywords: CRC with age; KRAS; RAS signalling; RAS-driven metastasis; cancer stem cells; colorectal cancer.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
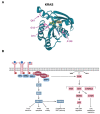
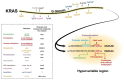

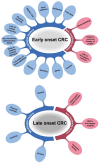
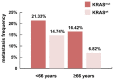
References
-
- Goswami D., Chen D., Yang Y., Gudla P., Columbus J., Worthy K., Rigby M., Wheeler M., Mukhopadhyay S., Powell K., et al. Membrane interactions of the globular domain and the hypervariable region of KRAS4b define its unique diffusion behavior. Elife. 2020;9 doi: 10.7554/eLife.47654. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

